2J1N
 
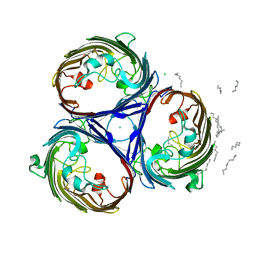 | | osmoporin OmpC | | Descriptor: | CHLORIDE ION, DODECANE, MAGNESIUM ION, ... | | Authors: | Basle, A, Storici, P, Rummel, G, Rosenbusch, J.P, Schirmer, T. | | Deposit date: | 2006-08-15 | | Release date: | 2006-09-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Osmoporin Ompc from E. Coli at 2.0 A.
J.Mol.Biol., 362, 2006
|
|
5OMT
 
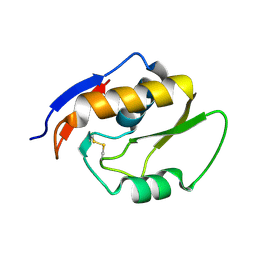 | | Endonuclease NucB | | Descriptor: | NucB | | Authors: | Basle, A, Lewis, R.J. | | Deposit date: | 2017-08-01 | | Release date: | 2017-11-15 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of NucB, a biofilm-degrading endonuclease.
Nucleic Acids Res., 46, 2018
|
|
5NQN
 
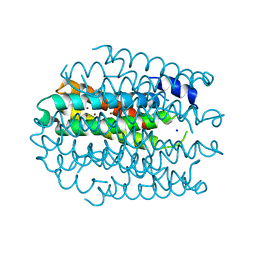 | |
5NQO
 
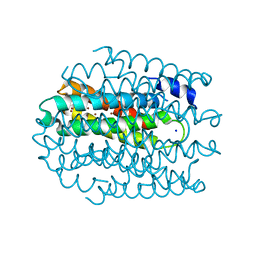 | |
5NQM
 
 | |
6Q64
 
 | | BT1044SeMet E190Q | | Descriptor: | Endoglycosidase | | Authors: | Basle, A, Paterson, N, Crouch, L, Bolam, D. | | Deposit date: | 2018-12-10 | | Release date: | 2019-05-08 | | Last modified: | 2019-09-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Complex N-glycan breakdown by gut Bacteroides involves an extensive enzymatic apparatus encoded by multiple co-regulated genetic loci.
Nat Microbiol, 4, 2019
|
|
6HZE
 
 | | BP0997, GH138 enzyme targeting pectin rhamnogalacturonan II | | Descriptor: | BPa0997, SODIUM ION, beta-D-galactopyranuronic acid | | Authors: | Basle, A, Cartmell, A, Labourel, A, Gilbert, H. | | Deposit date: | 2018-10-23 | | Release date: | 2019-03-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and functional analyses of glycoside hydrolase 138 enzymes targeting chain A galacturonic acid in the complex pectin rhamnogalacturonan II.
J.Biol.Chem., 294, 2019
|
|
6HZG
 
 | | BP0997, GH138 enzyme targeting pectin rhamnogalacturonan II | | Descriptor: | BPa0997 N-ter E361S, CHLORIDE ION, SODIUM ION, ... | | Authors: | Basle, A, Cartmell, A, Labourel, A, Gilbert, H. | | Deposit date: | 2018-10-23 | | Release date: | 2019-03-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional analyses of glycoside hydrolase 138 enzymes targeting chain A galacturonic acid in the complex pectin rhamnogalacturonan II.
J.Biol.Chem., 294, 2019
|
|
7ZNR
 
 | |
7ZNS
 
 | |
8C46
 
 | |
7NWR
 
 | | Structure of BT1526, a myo-inositol-1-phosphate synthase | | Descriptor: | Inositol-3-phosphate synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION | | Authors: | Basle, A, Tang, G, Marles-Wright, J, Campopiano, D. | | Deposit date: | 2021-03-17 | | Release date: | 2022-03-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of inositol lipid metabolism in gut-associated Bacteroidetes.
Nat Microbiol, 7, 2022
|
|
8AVN
 
 | | Mutant of Superoxide dismutase SodFM1 from CPR Parcubacteria Wolfebacteria | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase | | Authors: | Basle, A, Barwinska-Sendra, A, Sendra, K.M, Waldron, K. | | Deposit date: | 2022-08-26 | | Release date: | 2023-04-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | An ancient metalloenzyme evolves through metal preference modulation.
Nat Ecol Evol, 7, 2023
|
|
8AVK
 
 | | Superoxide dismutase SodFM1 from CPR Parkubacteria Wolfebacteria | | Descriptor: | FE (III) ION, Superoxide dismutase | | Authors: | Basle, A, Barwinska-Sendra, A, Sendra, K.M, Waldron, K. | | Deposit date: | 2022-08-26 | | Release date: | 2023-04-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An ancient metalloenzyme evolves through metal preference modulation.
Nat Ecol Evol, 7, 2023
|
|
8AVL
 
 | | Superoxide dismutase SodFM2 from Bacteroides fragilis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FE (III) ION, Superoxide dismutase [Fe] | | Authors: | Basle, A, Barwinska-Sendra, A, Sendra, K.M, Waldron, K. | | Deposit date: | 2022-08-26 | | Release date: | 2023-04-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An ancient metalloenzyme evolves through metal preference modulation.
Nat Ecol Evol, 7, 2023
|
|
8AVM
 
 | | Mutant of Superoxide Dismutase sodfm2 from Bacteroides fragilis | | Descriptor: | FE (III) ION, Superoxide dismutase [Fe] | | Authors: | Basle, A, Barwinska-Sendra, A, Sendra, K.M, Waldron, K. | | Deposit date: | 2022-08-26 | | Release date: | 2023-04-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An ancient metalloenzyme evolves through metal preference modulation.
Nat Ecol Evol, 7, 2023
|
|
7POA
 
 | | An Irreversible, Promiscuous and Highly Thermostable Claisen-Condensation Biocatalyst Drives the Synthesis of Substituted Pyrroles | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 8-amino-7-oxononanoate synthase/2-amino-3-ketobutyrate coenzyme A ligase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Basle, A, Ashley, B, Campopiano, D, Marles-Wright, J. | | Deposit date: | 2021-09-08 | | Release date: | 2022-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Versatile Chemo-Biocatalytic Cascade Driven by a Thermophilic and Irreversible C-C Bond-Forming alpha-Oxoamine Synthase.
Acs Sustain Chem Eng, 11, 2023
|
|
7POC
 
 | | An Irreversible, Promiscuous and Highly Thermostable Claisen-Condensation Biocatalyst Drives the Synthesis of Substituted Pyrroles | | Descriptor: | 8-amino-7-oxononanoate synthase/2-amino-3-ketobutyrate coenzyme A ligase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Basle, A, Ashley, B, Campopiano, D, Marles-Wright, J. | | Deposit date: | 2021-09-08 | | Release date: | 2022-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Versatile Chemo-Biocatalytic Cascade Driven by a Thermophilic and Irreversible C-C Bond-Forming alpha-Oxoamine Synthase.
Acs Sustain Chem Eng, 11, 2023
|
|
7POB
 
 | | An Irreversible, Promiscuous and Highly Thermostable Claisen-Condensation Biocatalyst Drives the Synthesis of Substituted Pyrroles | | Descriptor: | 8-amino-7-oxononanoate synthase/2-amino-3-ketobutyrate coenzyme A ligase, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION | | Authors: | Basle, A, Ashley, B, Campopiano, D, Marles-Wright, J. | | Deposit date: | 2021-09-08 | | Release date: | 2022-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Versatile Chemo-Biocatalytic Cascade Driven by a Thermophilic and Irreversible C-C Bond-Forming alpha-Oxoamine Synthase.
Acs Sustain Chem Eng, 11, 2023
|
|
5LA0
 
 | | The mechanism by which arabinoxylanases can recognise highly decorated xylans | | Descriptor: | CALCIUM ION, Carbohydrate binding family 6, SULFATE ION, ... | | Authors: | Basle, A, Labourel, A, Cuskin, F, Jackson, A, Crouch, L, Rogowski, A, Gilbert, A. | | Deposit date: | 2016-06-13 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Mechanism by Which Arabinoxylanases Can Recognize Highly Decorated Xylans.
J.Biol.Chem., 291, 2016
|
|
5LA1
 
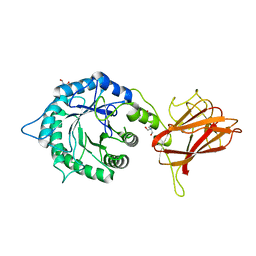 | | The mechanism by which arabinoxylanases can recognise highly decorated xylans | | Descriptor: | CALCIUM ION, Carbohydrate binding family 6, TRIS-HYDROXYMETHYL-METHYL-AMMONIUM, ... | | Authors: | Basle, A, Labourel, A, Cuskin, F, Jackson, A, Crouch, L, Rogowski, A, Gilbert, H. | | Deposit date: | 2016-06-13 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Mechanism by Which Arabinoxylanases Can Recognize Highly Decorated Xylans.
J.Biol.Chem., 291, 2016
|
|
5LA2
 
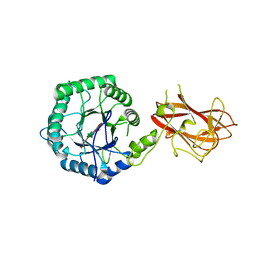 | | The mechanism by which arabinoxylanases can recognise highly decorated xylans | | Descriptor: | CALCIUM ION, Carbohydrate binding family 6, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-[alpha-L-arabinofuranose-(1-3)]alpha-D-xylopyranose, ... | | Authors: | Basle, A, Labourel, A, Cuskin, F, Jackson, A, Crouch, L, Rogowski, A, Gilbert, H. | | Deposit date: | 2016-06-13 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Mechanism by Which Arabinoxylanases Can Recognize Highly Decorated Xylans.
J.Biol.Chem., 291, 2016
|
|
5MSY
 
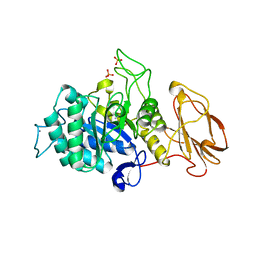 | | Glycoside hydrolase BT_1012 | | Descriptor: | AMMONIA, Glycoside hydrolase, PHOSPHATE ION | | Authors: | Basle, A, Ndeh, D, Rogowski, A, Cartmell, A, Luis, A.S, Venditto, I, Labourel, A, Gilbert, H.J. | | Deposit date: | 2017-01-06 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Complex pectin metabolism by gut bacteria reveals novel catalytic functions.
Nature, 544, 2017
|
|
5MQO
 
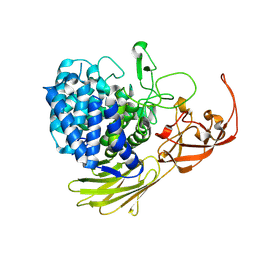 | | Glycoside hydrolase BT_1003 | | Descriptor: | Non-reducing end beta-L-arabinofuranosidase | | Authors: | Basle, A, Ndeh, D, Rogowski, A, Cartmell, A, Luis, A.S, Venditto, I, Labourel, A, Gilbert, H.J. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Complex pectin metabolism by gut bacteria reveals novel catalytic functions.
Nature, 544, 2017
|
|
5MQS
 
 | | Sialidase BT_1020 | | Descriptor: | Beta-L-arabinobiosidase, CALCIUM ION, SODIUM ION, ... | | Authors: | Basle, A, Ndeh, D, Rogowski, A, Cartmell, A, Luis, A.S, Venditto, I, Labourel, A, Gilbert, H.J. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Complex pectin metabolism by gut bacteria reveals novel catalytic functions.
Nature, 544, 2017
|
|
