3EE7
 
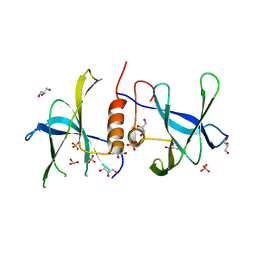 | | Crystal Structure of SARS-CoV nsp9 G104E | | Descriptor: | GLYCEROL, PHOSPHATE ION, Replicase polyprotein 1a | | Authors: | Miknis, Z.J, Donaldson, E.F, Umland, T.C, Rimmer, R, Baric, R.S, Schultz, L.W. | | Deposit date: | 2008-09-04 | | Release date: | 2009-03-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Severe acute respiratory syndrome coronavirus nsp9 dimerization is essential for efficient viral growth
J.Virol., 83, 2009
|
|
5H37
 
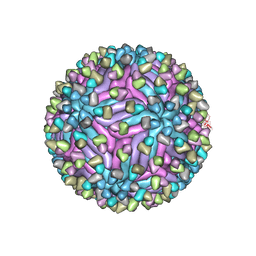 | | Cryo-EM structure of zika virus complexed with Fab C10 at pH 8.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C10 IgG heavy chain variable region, C10 IgG light chain variable region, ... | | Authors: | Zhang, S, Kostyuchenko, V, Ng, T.-S, Lim, X.-N, Ooi, J.S.G, Lambert, S, Tan, T.Y, Widman, D, Shi, J, Baric, R.S, Lok, S.-M. | | Deposit date: | 2016-10-20 | | Release date: | 2016-11-30 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Neutralization mechanism of a highly potent antibody against Zika virus
Nat Commun, 7, 2016
|
|
4L3N
 
 | | Crystal structure of the receptor-binding domain from newly emerged Middle East respiratory syndrome coronavirus | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S protein | | Authors: | Chen, Y, Rajashankar, K.R, Yang, Y, Agnihothram, S.S, Liu, C, Lin, Y.-L, Baric, R.S, Li, F. | | Deposit date: | 2013-06-06 | | Release date: | 2013-07-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structure of the receptor-binding domain from newly emerged middle East respiratory syndrome coronavirus.
J.Virol., 87, 2013
|
|
6XRZ
 
 | | The 28-kDa Frameshift Stimulation Element from the SARS-CoV-2 RNA Genome | | Descriptor: | Frameshift Stimulation Element from the SARS-CoV-2 RNA Genome | | Authors: | Zhang, K, Zheludev, I, Hagey, R, Wu, M, Haslecker, R, Hou, Y, Kretsch, R, Pintilie, G, Rangan, R, Kladwang, W, Li, S, Pham, E, Souibgui, C, Baric, R, Sheahan, T, Souza, V, Glenn, J, Chiu, W, Das, R. | | Deposit date: | 2020-07-14 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Cryo-electron Microscopy and Exploratory Antisense Targeting of the 28-kDa Frameshift Stimulation Element from the SARS-CoV-2 RNA Genome.
Biorxiv, 2020
|
|
8DI5
 
 | | Cryo-EM structure of SARS-CoV-2 Beta (B.1.351) spike protein in complex with VH domain F6 (focused refinement of RBD and VH F6) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, VH F6 | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Subramaniam, S. | | Deposit date: | 2022-06-28 | | Release date: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Potent and broad neutralization of SARS-CoV-2 variants of concern (VOCs) including omicron sub-lineages BA.1 and BA.2 by biparatopic human VH domains.
Iscience, 25, 2022
|
|
6N81
 
 | |
8SAK
 
 | | BtCoV-422 in complex with neutralizing antibody JC57-11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | McFadden, E, McLellan, J.S. | | Deposit date: | 2023-04-01 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A MERS-CoV antibody neutralizes a pre-emerging group 2c bat coronavirus.
Sci Transl Med, 15, 2023
|
|
6N8D
 
 | |
5H32
 
 | | Cryo-EM structure of zika virus complexed with Fab C10 at pH 5.0 | | Descriptor: | C10 IgG heavy chain variable region, C10 IgG light chain variable region, structural protein E | | Authors: | Zhang, S, Kostyuchenko, V, Ng, T.-S, Lok, S.-M. | | Deposit date: | 2016-10-20 | | Release date: | 2016-11-30 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Neutralization mechanism of a highly potent antibody against Zika virus
Nat Commun, 7, 2016
|
|
5H30
 
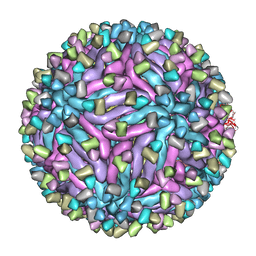 | | Cryo-EM structure of zika virus complexed with Fab C10 at pH 6.5 | | Descriptor: | IgG C10 heavy chain, IgG C10 light chain, structural protein E, ... | | Authors: | Zhang, S, Kostyuchenko, V, Ng, T.-S, Lok, S.-M. | | Deposit date: | 2016-10-19 | | Release date: | 2016-11-30 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Neutralization mechanism of a highly potent antibody against Zika virus
Nat Commun, 7, 2016
|
|
7LJR
 
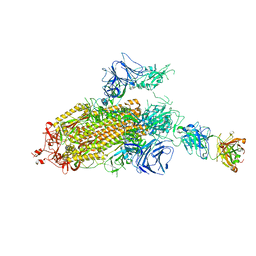 | | SARS-CoV-2 Spike Protein Trimer bound to DH1043 fab | | Descriptor: | Fab DH1043 heavy chain, Fab DH1043 light chain, Spike glycoprotein | | Authors: | Gobeil, S, Acharya, P. | | Deposit date: | 2021-01-30 | | Release date: | 2021-03-31 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | The functions of SARS-CoV-2 neutralizing and infection-enhancing antibodies in vitro and in mice and nonhuman primates.
Biorxiv, 2021
|
|
7M8J
 
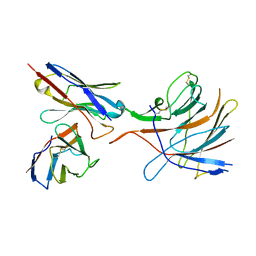 | | SARS-CoV-2 S-NTD + Fab CM25 | | Descriptor: | CM25 Fab - Heavy Chain, CM25 Fab - Light Chain, Spike protein S1 | | Authors: | Johnson, N.V, Mclellan, J.S. | | Deposit date: | 2021-03-29 | | Release date: | 2021-05-19 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Prevalent, protective, and convergent IgG recognition of SARS-CoV-2 non-RBD spike epitopes.
Science, 372, 2021
|
|
7LRT
 
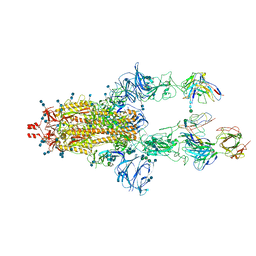 | |
7LRS
 
 | |
7MLZ
 
 | | Cryo-EM structure of SARS-CoV-2 spike in complex with neutralizing antibody B1-182.1 that targets the receptor-binding domain | | Descriptor: | B1-182.1 Fab heavy chain, B1-182.1 Fab light chain, Spike protein S1, ... | | Authors: | Zhou, T, Tsybovsky, T, Kwong, P.D. | | Deposit date: | 2021-04-29 | | Release date: | 2021-07-28 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Ultrapotent antibodies against diverse and highly transmissible SARS-CoV-2 variants.
Science, 373, 2021
|
|
7MM0
 
 | | Cryo-EM structure of SARS-CoV-2 spike in complex with neutralizing antibody B1-182.1 that targets the receptor-binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, B1-182.1 Fab heavy chain, ... | | Authors: | Zhou, T, Tsybovsky, T, Kwong, P.D. | | Deposit date: | 2021-04-29 | | Release date: | 2021-07-28 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Ultrapotent antibodies against diverse and highly transmissible SARS-CoV-2 variants.
Science, 373, 2021
|
|
7S2S
 
 | | nanobody bound to Interleukin-2Rbeta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, IL2Rb-binding nanobody, Interleukin-2 receptor subunit beta, ... | | Authors: | Glassman, C.R, Jude, K.M, Yen, M, Garcia, K.C. | | Deposit date: | 2021-09-03 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Facile discovery of surrogate cytokine agonists.
Cell, 185, 2022
|
|
7S2R
 
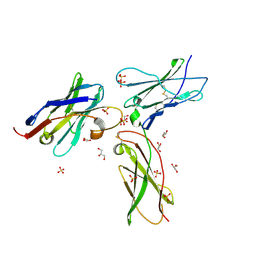 | | nanobody bound to IL-2Rg | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokine receptor common subunit gamma, ... | | Authors: | Glassman, C.R, Jude, K.M, Yen, M, Garcia, K.C. | | Deposit date: | 2021-09-03 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Facile discovery of surrogate cytokine agonists.
Cell, 185, 2022
|
|
7S3N
 
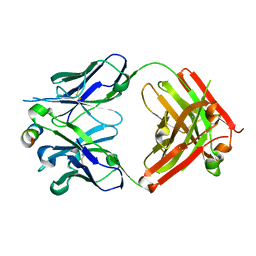 | |
7S3M
 
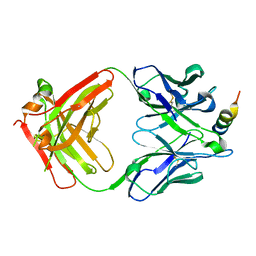 | |
7SG4
 
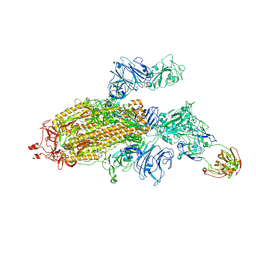 | |
7T3M
 
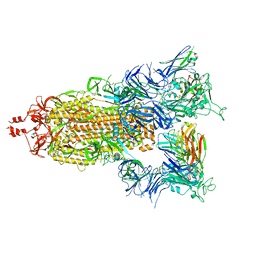 | | SARS-CoV-2 S (Spike Glycoprotein) D614G with Three (3) RBDs Up, Bound to Antibody 2-7 scFv, composite map | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody 2-7 scFv, ... | | Authors: | Byrne, P.O, McLellan, J.S. | | Deposit date: | 2021-12-08 | | Release date: | 2022-08-24 | | Last modified: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | IgG-like bispecific antibodies with potent and synergistic neutralization against circulating SARS-CoV-2 variants of concern.
Nat Commun, 13, 2022
|
|
7T67
 
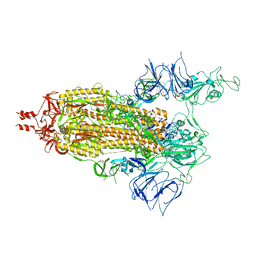 | | SARS-CoV-2 S (Spike Glycoprotein) D614G with One(1) RBD Up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Byrne, P.O, McLellan, J.S. | | Deposit date: | 2021-12-13 | | Release date: | 2022-08-24 | | Last modified: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | IgG-like bispecific antibodies with potent and synergistic neutralization against circulating SARS-CoV-2 variants of concern.
Nat Commun, 13, 2022
|
|
7TB4
 
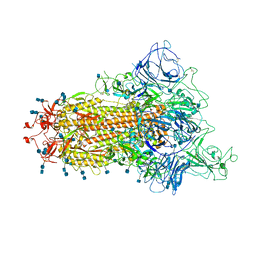 | | Cryo-EM structure of the spike of SARS-CoV-2 Omicron variant of concern | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Surface glycoprotein | | Authors: | Zhou, T, Tsybovsky, T, Kwong, P.D. | | Deposit date: | 2021-12-21 | | Release date: | 2022-01-12 | | Last modified: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Antibodies with potent and broad neutralizing activity against antigenically diverse and highly transmissible SARS-CoV-2 variants.
Biorxiv, 2021
|
|
8DGU
 
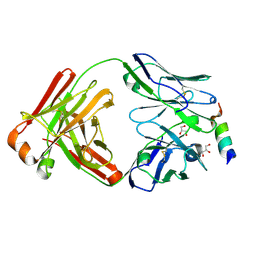 | |
