6GV9
 
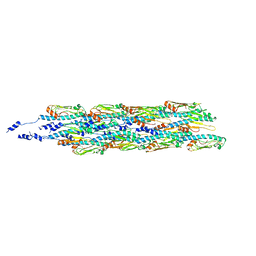 | | Structure of the type IV pilus from enterohemorrhagic Escherichia coli (EHEC) | | Descriptor: | Prepilin peptidase-dependent protein D | | Authors: | Bardiaux, B, Amorim, G.C, Luna-Rico, A, Zheng, W, Guilvout, I, Jollivet, C, Nilges, M, Egelman, E, Francetic, O, Izadi-Pruneyre, N. | | Deposit date: | 2018-06-20 | | Release date: | 2019-05-15 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structure and Assembly of the Enterohemorrhagic Escherichia coli Type 4 Pilus.
Structure, 27, 2019
|
|
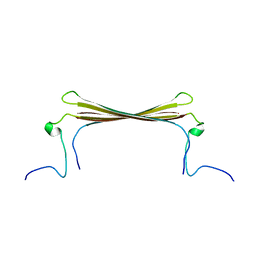
6RHY
 
 | |
6GMS
 
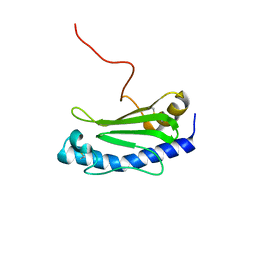 | | Solution NMR structure of the major type IV pilin PpdD from enterohemorrhagic Escherichia coli (EHEC) | | Descriptor: | Prepilin peptidase-dependent protein D | | Authors: | Amorim, G.C, Bardiaux, B, Luna-Rico, A, Zeng, W, Guilvout, I, Egelman, E, Nilges, M, Francetic, O, Izadi-Pruneyre, N. | | Deposit date: | 2018-05-28 | | Release date: | 2019-05-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure and Assembly of the Enterohemorrhagic Escherichia coli Type 4 Pilus.
Structure, 27, 2019
|
|
3J07
 
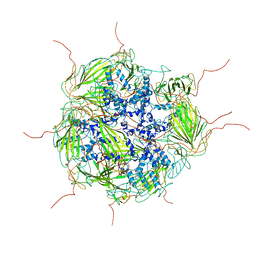 | | Model of a 24mer alphaB-crystallin multimer | | Descriptor: | Alpha-crystallin B chain | | Authors: | Jehle, S, Vollmar, B, Bardiaux, B, Dove, K.K, Rajagopal, P, Gonen, T, Oschkinat, H, Klevit, R.E. | | Deposit date: | 2011-04-27 | | Release date: | 2016-01-20 | | Last modified: | 2019-04-24 | | Method: | ELECTRON MICROSCOPY (20 Å), SOLID-STATE NMR, SOLUTION SCATTERING | | Cite: | N-terminal domain of {alpha}B-crystallin provides a conformational switch for multimerization and structural heterogeneity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5WDA
 
 | | Structure of the PulG pseudopilus | | Descriptor: | CALCIUM ION, General secretion pathway protein G | | Authors: | Lopez-Castilla, A, Thomassin, J.L, Bardiaux, B, Zheng, W, Nivaskumar, M, Yu, X, Nilges, M, Egelman, E.H, Izadi-Pruneyre, N, Francetic, O. | | Deposit date: | 2017-07-04 | | Release date: | 2017-10-25 | | Last modified: | 2020-01-01 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structure of the calcium-dependent type 2 secretion pseudopilus.
Nat Microbiol, 2, 2017
|
|
6YP5
 
 | |
8A9W
 
 | | Crystal structure of PulL C-ter domain | | Descriptor: | SULFATE ION, Type II secretion system protein L | | Authors: | Dazzoni, R, Li, Y, Lopez-Castilla, A, Brier, S, Mechaly, A, Cordier, F, Haouz, A, Nilges, M, Francetic, O, Bardiaux, B, Izadi-Pruneyre, N. | | Deposit date: | 2022-06-29 | | Release date: | 2023-01-11 | | Last modified: | 2023-04-12 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Structure and dynamic association of an assembly platform subcomplex of the bacterial type II secretion system.
Structure, 31, 2023
|
|
8A9X
 
 | | Crystal structure of PulM C-ter domain | | Descriptor: | Type II secretion system protein M | | Authors: | Dazzoni, R, Li, Y, Lopez-Castilla, A, Brier, S, Mechaly, A, Cordier, F, Haouz, A, Nilges, M, Francetic, O, Bardiaux, B, Izadi-Pruneyre, N. | | Deposit date: | 2022-06-29 | | Release date: | 2023-01-25 | | Last modified: | 2023-04-12 | | Method: | X-RAY DIFFRACTION (1.523 Å) | | Cite: | Structure and dynamic association of an assembly platform subcomplex of the bacterial type II secretion system.
Structure, 31, 2023
|
|
8PEK
 
 | |
6EKA
 
 | | Solid-state MAS NMR structure of the HELLF prion amyloid fibrils | | Descriptor: | Podospora anserina S mat+ genomic DNA chromosome 3, supercontig 2 | | Authors: | Martinez, D, Daskalov, A, Andreas, L, Bardiaux, B, Coustou, V, Stanek, J, Berbon, M, Noubhani, M, Kauffmann, B, Wall, J.S, Pintacuda, G, Saupe, S.J, Habenstein, B, Loquet, A. | | Deposit date: | 2017-09-25 | | Release date: | 2018-10-10 | | Last modified: | 2023-06-14 | | Method: | SOLID-STATE NMR | | Cite: | Structural and molecular basis of cross-seeding barriers in amyloids
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6G81
 
 | | Solution structure of the Ni metallochaperone HypA from Helicobacter pylori | | Descriptor: | Hydrogenase maturation factor HypA, ZINC ION | | Authors: | Spronk, C.A.E.M, Zerko, S, Gorka, M, Kozminski, W, Bardiaux, B, Zambelli, B, Musiani, F, Piccioli, M, Hu, H, Maroney, M, Ciurli, S. | | Deposit date: | 2018-04-07 | | Release date: | 2018-10-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of Helicobacter pylori nickel-chaperone HypA: an integrated approach using NMR spectroscopy, functional assays and computational tools.
J. Biol. Inorg. Chem., 23, 2018
|
|
5O2Y
 
 | | NMR structure of the calcium bound form of PulG, major pseudopilin from Klebsiella oxytoca T2SS | | Descriptor: | CALCIUM ION, General secretion pathway protein G | | Authors: | Lopez-Castilla, A, Bardiaux, B, Vitorge, B, Thomassin, J.-L, Zheng, W, Yu, X, Egelman, E.H, Nilges, M, Francetic, O, Izadi-Pruneyre, N. | | Deposit date: | 2017-05-23 | | Release date: | 2017-10-18 | | Last modified: | 2019-05-08 | | Method: | SOLUTION NMR | | Cite: | Structure of the calcium-dependent type 2 secretion pseudopilus.
Nat Microbiol, 2, 2017
|
|
5LME
 
 | | Specific-DNA binding activity of the cross-brace zinc finger motif of the piggyBac transposase | | Descriptor: | ZINC ION, piggyBac transposase | | Authors: | Morellet, N, Taylor, J.A, Wieninger, S, Moriau, S, Li, X, Lescop, E, Mathy, N, Bischerour, J, Betermier, M, Bardiaux, B, Nilges, M, Craig, N.L, Hickman, A.B, Dyda, F, Guittet, E. | | Deposit date: | 2016-07-30 | | Release date: | 2017-12-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Sequence-specific DNA binding activity of the cross-brace zinc finger motif of the piggyBac transposase.
Nucleic Acids Res., 46, 2018
|
|
5MWV
 
 | | Solid-state NMR Structure of outer membrane protein G in lipid bilayers | | Descriptor: | Outer membrane protein G | | Authors: | Retel, J.S, Nieuwkoop, A.J, Hiller, M, Higman, V.A, Barbet-Massin, E, Stanek, J, Andreas, L.B, Franks, W.T, van Rossum, B.-J, Vinothkumar, K.R, Handel, L, de Palma, G.G, Bardiaux, B, Pintacuda, G, Emsley, L, Kuelbrandt, W, Oschkinat, H. | | Deposit date: | 2017-01-20 | | Release date: | 2017-12-27 | | Last modified: | 2019-08-21 | | Method: | SOLID-STATE NMR | | Cite: | Structure of outer membrane protein G in lipid bilayers.
Nat Commun, 8, 2017
|
|
7ZBQ
 
 | | Structure of the ADP-ribosyltransferase TccC3HVR from Photorhabdus Luminescens | | Descriptor: | TccC3 | | Authors: | Lindemann, F, Belyy, A, Friedrich, D, Schmieder, P, Bardiaux, B, Roderer, D, Funk, J, Protze, J, Bieling, P, Oschkinat, H, Raunser, S. | | Deposit date: | 2022-03-24 | | Release date: | 2022-06-29 | | Last modified: | 2022-08-03 | | Method: | SOLUTION NMR | | Cite: | Mechanism of threonine ADP-ribosylation of F-actin by a Tc toxin.
Nat Commun, 13, 2022
|
|
7ZE0
 
 | | Solution structure of the PulM C-terminal domain from Klebsiella oxytoca | | Descriptor: | Type II secretion system protein M | | Authors: | Lopez-Castilla, A, Bardiaux, B, Nilges, M, Francetic, O, Izadi-Pruneyre, N. | | Deposit date: | 2022-03-30 | | Release date: | 2023-01-18 | | Last modified: | 2023-02-15 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamic association of an assembly platform subcomplex of the bacterial type II secretion system.
Structure, 31, 2023
|
|
8AB1
 
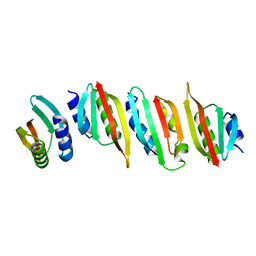 | | Crystal structure of the PulL-PulM C-terminal domain heterocomplex | | Descriptor: | Type II secretion system protein L, Type II secretion system protein M | | Authors: | Dazzoni, R, Li, Y, Lopez-Castilla, A, Brier, S, Mechaly, A, Cordier, F, Haouz, A, Nilges, M, Francetic, O, Bardiaux, B, Izadi-Pruneyre, N. | | Deposit date: | 2022-07-04 | | Release date: | 2023-01-11 | | Last modified: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structure and dynamic association of an assembly platform subcomplex of the bacterial type II secretion system.
Structure, 31, 2023
|
|
2MWG
 
 | |
2RLZ
 
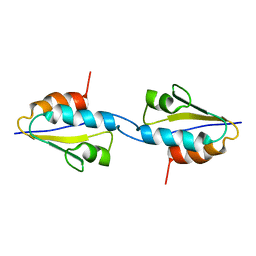 | | Solid-State MAS NMR structure of the dimer Crh | | Descriptor: | HPr-like protein crh | | Authors: | Loquet, A, Bardiaux, B, Gardiennet, C, Blanchet, C, Baldus, M, Nilges, M, Malliavin, T, Bockmann, A. | | Deposit date: | 2007-09-04 | | Release date: | 2008-06-17 | | Last modified: | 2022-03-16 | | Method: | SOLID-STATE NMR | | Cite: | 3D structure determination of the Crh protein from highly ambiguous solid-state NMR restraints.
J.Am.Chem.Soc., 130, 2008
|
|
2KLR
 
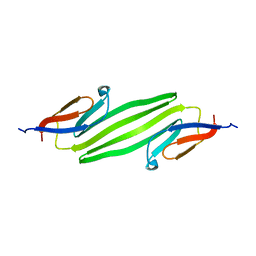 | | Solid-state NMR structure of the alpha-crystallin domain in alphaB-crystallin oligomers | | Descriptor: | Alpha-crystallin B chain | | Authors: | Jehle, S, Rajagopal, P, Markovic, S, Bardiaux, B, Kuehne, R, Higman, V.A, Klevit, R.E, van Rossum, B, Oschkinat, H. | | Deposit date: | 2009-07-08 | | Release date: | 2010-07-07 | | Last modified: | 2011-07-13 | | Method: | SOLID-STATE NMR | | Cite: | Solid-state NMR and SAXS studies provide a structural basis for the activation of alphaB-crystallin oligomers.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2LME
 
 | | Solid-state NMR structure of the membrane anchor domain of the trimeric autotransporter YadA | | Descriptor: | Adhesin yadA | | Authors: | Shahid, S.A, Bardiaux, B, Franks, W.T, Habeck, M, Linke, D, van Rossum, B. | | Deposit date: | 2011-11-30 | | Release date: | 2012-11-07 | | Last modified: | 2023-06-14 | | Method: | SOLID-STATE NMR | | Cite: | Membrane-protein structure determination by solid-state NMR spectroscopy of microcrystals.
Nat.Methods, 9, 2012
|
|
2KUB
 
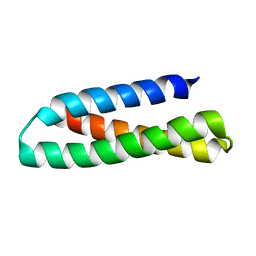 | | Solution structure of the alpha subdomain of the major non-repeat unit of Fap1 fimbriae of Streptococcus parasanguis | | Descriptor: | Fimbriae-associated protein Fap1 | | Authors: | Ramboarina, S, Garnett, J.A, Bodey, A, Simpson, P, Bardiaux, B, Nilges, M, Matthews, S. | | Deposit date: | 2010-02-17 | | Release date: | 2010-07-21 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structural insights into serine-rich fimbriae from gram-positive bacteria.
J.Biol.Chem., 2010
|
|
2MS7
 
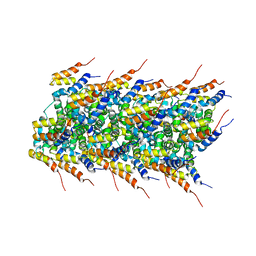 | | High-resolution solid-state NMR structure of the helical signal transduction filament MAVS CARD | | Descriptor: | Mitochondrial antiviral-signaling protein | | Authors: | He, L, Bardiaux, B, Spehr, J, Luehrs, T, Ritter, C. | | Deposit date: | 2014-07-25 | | Release date: | 2015-09-02 | | Last modified: | 2016-02-03 | | Method: | SOLID-STATE NMR | | Cite: | Structure determination of helical filaments by solid-state NMR spectroscopy.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2X12
 
 | |
8P9R
 
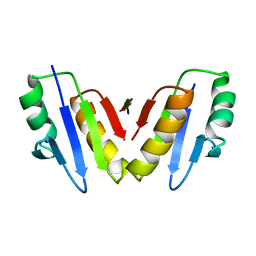 | | Structure of the periplasmic domain of ExbD from E. coli in complex with TonB | | Descriptor: | Biopolymer transport protein ExbD, Protein TonB | | Authors: | Zinke, M, Mechaly, A, Lejeune, M, Haouz, A, Izadi-Pruneyre, N. | | Deposit date: | 2023-06-06 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Ton Motor Conformational Switch and Peptidoglycan Role in Bacterial Nutrient Uptake.
Biorxiv, 2023
|
|
