4E2D
 
 | | Structure of the old yellow enzyme from Trypanosoma cruzi | | Descriptor: | DIMETHYL SULFOXIDE, FLAVIN MONONUCLEOTIDE, Old Yellow Protein | | Authors: | Murakami, M.T, Rodrigues, N.C, Gava, L.M, Canduri, F, Oliva, G, Barbosa, L.R.S, Borgers, J.C. | | Deposit date: | 2012-03-08 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | High resolution crystal structure and in solution studies of the Old Yellow Enzyme from Trypanosoma cruzi: Insights into oligomerization, enzyme dynamics and specificity
To be Published
|
|
4E2B
 
 | | High resolution crystal structure of the old yellow enzyme from Trypanosoma cruzi | | Descriptor: | DI(HYDROXYETHYL)ETHER, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Murakami, M.T, Rodrigues, N.C, Gava, L.M, Canduri, F, Oliva, G, Barbosa, L.R.S, Borgers, J.C. | | Deposit date: | 2012-03-08 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.269 Å) | | Cite: | High resolution crystal structure and in solution studies of the old yellow enzyme from Trypanosoma cruzi: Insights into oligomerization, enzyme dynamics and specificity
To be Published
|
|
3TH1
 
 | | Crystal structure of chlorocatechol 1,2-dioxygenase from Pseudomonas putida | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, ACETATE ION, Chlorocatechol 1,2-dioxygenase, ... | | Authors: | Rustiguel, J.K, Nonato, M.C. | | Deposit date: | 2011-08-18 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal Structure of chlorocatechol 1,2-dioxygenase from Pseudomonas putida
To be Published
|
|
6BBA
 
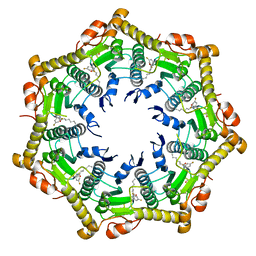 | |
6NB1
 
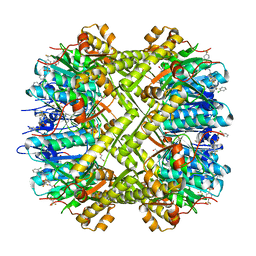 | | Crystal structure of Escherichia coli ClpP protease complexed with small molecule activator, ACP1-06 | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, GLYCEROL, N-{2-[(2-chlorophenyl)sulfanyl]ethyl}-2-methyl-2-{[5-(trifluoromethyl)pyridin-2-yl]sulfonyl}propanamide | | Authors: | Mabanglo, M.F, Houry, W.A, Eger, B.T, Bryson, S, Pai, E.F. | | Deposit date: | 2018-12-06 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | ClpP protease activation results from the reorganization of the electrostatic interaction networks at the entrance pores.
Commun Biol, 2, 2019
|
|
6NAY
 
 | |
6NAH
 
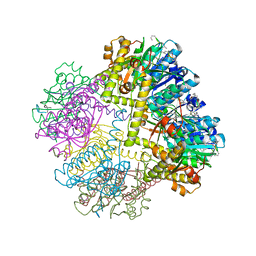 | |
6NAW
 
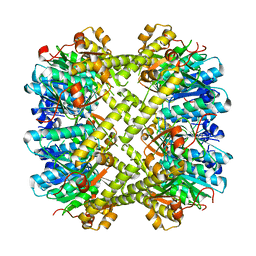 | |
6NAQ
 
 | | Crystal structure of Neisseria meningitidis ClpP protease in Apo form | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, POTASSIUM ION | | Authors: | Houry, W.A, Mabanglo, M.F, Pai, E.F, Eger, B.T, Bryson, S. | | Deposit date: | 2018-12-06 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.022 Å) | | Cite: | ClpP protease activation results from the reorganization of the electrostatic interaction networks at the entrance pores.
Commun Biol, 2, 2019
|
|
