4Y2O
 
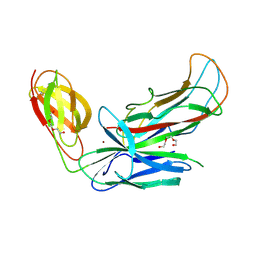 | | Structure of CFA/I pili chaperone-major subunit complex (CfaA-CfaB) | | Descriptor: | CFA/I fimbrial subunit B, CfA/I fimbrial subunit A (Colonization factor antigen subunit A putative chaperone), NICKEL (II) ION, ... | | Authors: | Bao, R, Xia, D. | | Deposit date: | 2015-02-10 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.419 Å) | | Cite: | Off-pathway assembly of fimbria subunits is prevented by chaperone CfaA of CFA/I fimbriae from enterotoxigenic E. coli.
Mol. Microbiol., 102, 2016
|
|
4Y2N
 
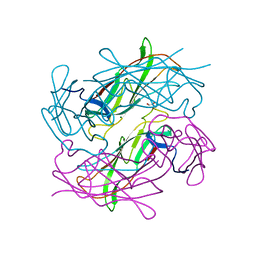 | | Structure of CFA/I pili major subunit CfaB trimer | | Descriptor: | 1,2-ETHANEDIOL, CFA/I fimbrial subunit B, MAGNESIUM ION | | Authors: | Bao, R, Xia, D. | | Deposit date: | 2015-02-10 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Off-pathway assembly of fimbria subunits is prevented by chaperone CfaA of CFA/I fimbriae from enterotoxigenic E. coli.
Mol. Microbiol., 102, 2016
|
|
4Y2L
 
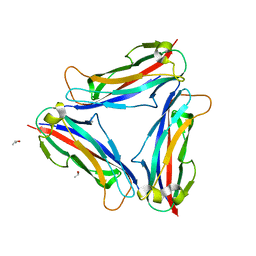 | | Structure of CFA/I pili major subunit CfaB trimer | | Descriptor: | CALCIUM ION, CFA/I fimbrial subunit B, ISOPROPYL ALCOHOL | | Authors: | Bao, R, Xia, D. | | Deposit date: | 2015-02-10 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.746 Å) | | Cite: | Off-pathway assembly of fimbria subunits is prevented by chaperone CfaA of CFA/I fimbriae from enterotoxigenic E. coli.
Mol. Microbiol., 102, 2016
|
|
4NCD
 
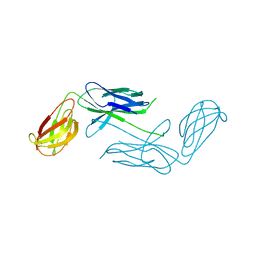 | | Crystal Structure of Class 5 Fimbriae Chaperone CfaA | | Descriptor: | Gram-negative pili assembly chaperone, N-terminal domain protein | | Authors: | Bao, R, Xia, D. | | Deposit date: | 2013-10-24 | | Release date: | 2014-08-27 | | Method: | X-RAY DIFFRACTION (2.037 Å) | | Cite: | Structure of CfaA Suggests a New Family of Chaperones Essential for Assembly of Class 5 Fimbriae.
Plos Pathog., 10, 2014
|
|
2FA4
 
 | |
2OE1
 
 | | Crystal Structure of Mitochondrial Thioredoxin 3 from Saccharomyces cerevisiae (reduced form) | | Descriptor: | SULFATE ION, Thioredoxin-3 | | Authors: | Bao, R, Zhang, Y.R, Zhou, C.Z, Chen, Y.X. | | Deposit date: | 2006-12-28 | | Release date: | 2008-01-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and mechanistic analyses of yeast mitochondrial thioredoxin Trx3 reveal putative function of its additional cysteine residues
Biochim.Biophys.Acta, 1794, 2009
|
|
2OE0
 
 | |
2OE3
 
 | |
3E8L
 
 | | The Crystal Structure of the Double-headed Arrowhead Protease Inhibitor A in Complex with Two Trypsins | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Bao, R, Jiang, C.-H, Chi, C.W, Lin, S.X, Chen, Y.X. | | Deposit date: | 2008-08-20 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | The ternary structure of double-headed arrowhead protease inhibitor API-A complexed with two trypsins reveals a novel reactive site conformation.
J.Biol.Chem., 284, 2009
|
|
4F8N
 
 | | X-ray structure of PsaA from Yersinia pestis, in complex with galactose and phosphate choline | | Descriptor: | CHLORIDE ION, GUANIDINE, PHOSPHOCHOLINE, ... | | Authors: | Bao, R, Esser, L, Xia, D. | | Deposit date: | 2012-05-17 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural basis for the specific recognition of dual receptors by the homopolymeric pH 6 antigen (Psa) fimbriae of Yersinia pestis.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4F8P
 
 | | X-ray structure of PsaA from Yersinia pestis, in complex with galactose | | Descriptor: | ACETATE ION, TERT-BUTYL FORMATE, beta-D-galactopyranose, ... | | Authors: | Bao, R, Esser, L, Xia, D. | | Deposit date: | 2012-05-17 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for the specific recognition of dual receptors by the homopolymeric pH 6 antigen (Psa) fimbriae of Yersinia pestis.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4F8O
 
 | | X-ray structure of PsaA from Yersinia pestis, in complex with lactose and AEBSF | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, CHLORIDE ION, GUANIDINE, ... | | Authors: | Bao, R, Esser, L, Xia, D. | | Deposit date: | 2012-05-17 | | Release date: | 2013-05-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the specific recognition of dual receptors by the homopolymeric pH 6 antigen (Psa) fimbriae of Yersinia pestis.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4F8L
 
 | | X-ray structure of PsaA from Yersinia pestis, in complex with galactose and AEBSF | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, BROMIDE ION, GLYCINE, ... | | Authors: | Bao, R, Esser, L, Xia, D. | | Deposit date: | 2012-05-17 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for the specific recognition of dual receptors by the homopolymeric pH 6 antigen (Psa) fimbriae of Yersinia pestis.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6IW7
 
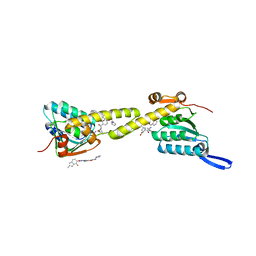 | | structural insights into Mycobacterium tuberculosis ClpP1P2 inhibition by Cediranib: implications for developing antimicrobial agents targeting Clp protease | | Descriptor: | (2S)-2-benzamido-4-methyl-pentanoic acid, 4-[(4-fluoro-2-methyl-3H-indol-5-yl)oxy]-6-methoxy-7-[3-(pyrrolidin-1-yl)propoxy]quinazoline, ATP-dependent Clp protease proteolytic subunit 1, ... | | Authors: | Bao, R, Luo, Y.F, Zhu, Y.B, Yang, Y, Zhou, Y.Z. | | Deposit date: | 2018-12-04 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.692121 Å) | | Cite: | structural insights into Mycobacterium tuberculosis ClpP1P2 inhibition by Cediranib: implications for developing antimicrobial agents targeting Clp protease
To Be Published
|
|
7W2O
 
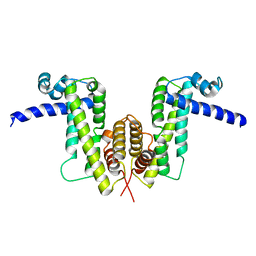 | |
7W2Q
 
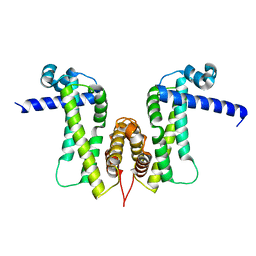 | |
5YFE
 
 | |
5WZE
 
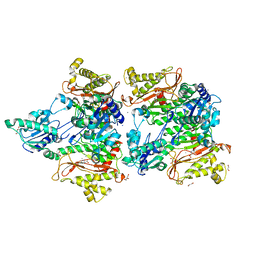 | | The structure of Pseudomonas aeruginosa aminopeptidase PepP | | Descriptor: | 1,2-ETHANEDIOL, ALANINE, Aminopeptidase P, ... | | Authors: | Bao, R, Peng, C.T, Liu, L, He, L.H, Li, C.C, Li, T, Shen, Y.L, Zhu, Y.B, Song, Y.J. | | Deposit date: | 2017-01-17 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | Structure-Function Relationship of Aminopeptidase P from Pseudomonas aeruginosa.
Front Microbiol, 8, 2017
|
|
7VK9
 
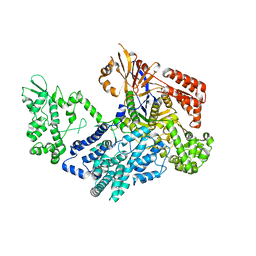 | | Crystal structure of xCas9 P411T | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1 | | Authors: | Bao, R, Liu, H.Y, Luo, Y.Z, Song, Y.J. | | Deposit date: | 2021-09-29 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and Dynamics Studies of the Spcas9 Variant Provide Insights into the Regulatory Role of the REC1 Domain
Acs Catalysis, 12, 2022
|
|
3F3R
 
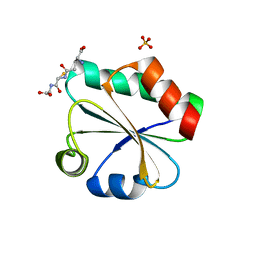 | | Crystal structure of yeast Thioredoxin1-glutathione mixed disulfide complex | | Descriptor: | GLUTATHIONE, SULFATE ION, Thioredoxin-1 | | Authors: | Zhang, Y.R, Bao, R, Zhou, C.Z, Chen, Y.X. | | Deposit date: | 2008-10-31 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and kinetic analysis of Saccharomyces cerevisiae thioredoxin Trx1: implications for the catalytic mechanism of GSSG reduced by the thioredoxin system
Biochim.Biophys.Acta, 1794, 2009
|
|
3F3Q
 
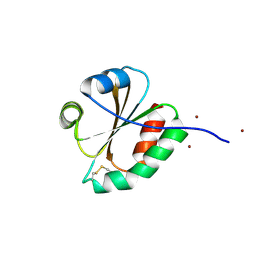 | | Crystal structure of the oxidised form of thioredoxin 1 from saccharomyces cerevisiae | | Descriptor: | Thioredoxin-1, ZINC ION | | Authors: | Zhang, Y.R, Bao, R, Zhou, C.Z, Chen, Y.X. | | Deposit date: | 2008-10-31 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural and kinetic analysis of Saccharomyces cerevisiae thioredoxin Trx1: implications for the catalytic mechanism of GSSG reduced by the thioredoxin system
Biochim.Biophys.Acta, 1794, 2009
|
|
7EZT
 
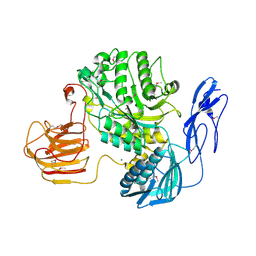 | | The structure and functional mechanism of nucleotide regulated acetylhexosaminidase Am2136 from Akkermansia muciniphila | | Descriptor: | Beta-N-acetylhexosaminidase, MAGNESIUM ION | | Authors: | Bao, R, Li, C.C, Tang, X.Y, Zhu, Y.B, Song, Y.J, Zhao, N.L, Huang, Q, Mou, X.Y, Luo, G.H, Liu, T.G. | | Deposit date: | 2021-06-02 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Nucleotide binding as an allosteric regulatory mechanism for Akkermansia muciniphila beta- N -acetylhexosaminidase Am2136.
Gut Microbes, 14, 2022
|
|
6K73
 
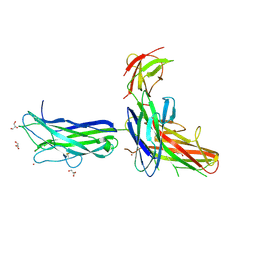 | |
7X8X
 
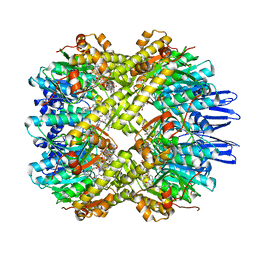 | | structural insights into Mycobacterium tuberculosis ClpP1P2 inhibition by Cediranib: implications for developing antimicrobial agents targeting Clp protease | | Descriptor: | 4-[1-[3-[4-[(4-fluoranyl-2-methyl-1H-indol-5-yl)oxy]-6-methoxy-quinazolin-7-yl]oxypropyl]piperidin-4-yl]benzamide, 4-[[3,5-bis(fluoranyl)phenyl]methyl]-N-[(4-bromophenyl)methyl]piperazine-1-carboxamide, ATP-dependent Clp protease proteolytic subunit 1, ... | | Authors: | Bao, R, Luo, Y.F, Zhu, Y.B, Yang, Y, Zhou, Y.Z. | | Deposit date: | 2022-03-15 | | Release date: | 2023-04-12 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Discovery and Mechanistic Study of Novel Mycobacterium tuberculosis ClpP1P2 Inhibitors
J.Med.Chem., 66, 2023
|
|
7EBQ
 
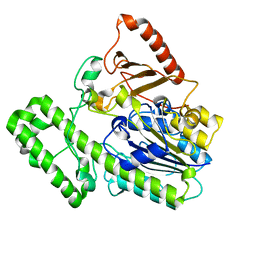 | | The structural analysis of A.Muciniphila sulfatase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Sulfatase | | Authors: | Bao, R, Li, C.C, Tang, X.Y, Zhu, Y.B, Song, Y.J, Zhao, N.L, Huang, Q, Mou, X.Y, Luo, G.H, Liu, T.G. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural analysis of the sulfatase AmAS from Akkermansia muciniphila.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
