6ID6
 
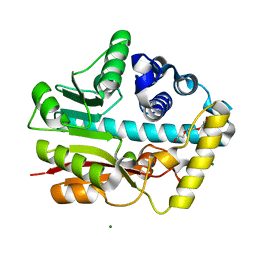 | |
5XC0
 
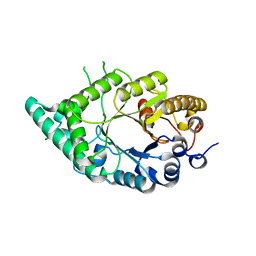 | |
5XC1
 
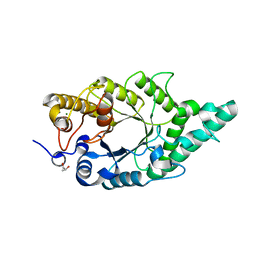 | | Crystal structure of the complex of an aromatic mutant (W6A) of an alkali thermostable GH10 Xylanase from Bacillus sp. NG-27 with S-1,2-Propanediol | | Descriptor: | Beta-xylanase, MAGNESIUM ION, S-1,2-PROPANEDIOL, ... | | Authors: | Bansia, H, Mahanta, P, Ramakumar, S. | | Deposit date: | 2017-03-21 | | Release date: | 2018-03-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Small Glycols Discover Cryptic Pockets on Proteins for Fragment-Based Approaches.
J.Chem.Inf.Model., 2021
|
|
5Z3J
 
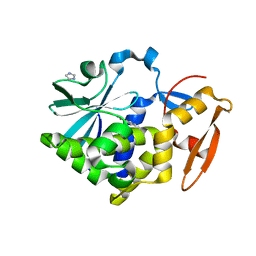 | |
5Z3I
 
 | |
5Z37
 
 | |
7ZQS
 
 | | Cryo-EM Structure of Human Transferrin Receptor 1 bound to DNA Aptamer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DNA (30-MER), ... | | Authors: | Bansia, H, Wang, T, Gutierrez, D, des Georges, A. | | Deposit date: | 2022-05-02 | | Release date: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Discovery of a Transferrin Receptor 1-Binding Aptamer and Its Application in Cancer Cell Depletion for Adoptive T-Cell Therapy Manufacturing.
J.Am.Chem.Soc., 144, 2022
|
|
5EFD
 
 | | Crystal structure of a surface pocket creating mutant (W6A) of an alkali thermostable GH10 xylanase from Bacillus sp. NG-27 | | Descriptor: | 1,2-ETHANEDIOL, Beta-xylanase, CHLORIDE ION, ... | | Authors: | Mahanta, P, Bhardwaj, A, Reddy, V.S, Ramakumar, S. | | Deposit date: | 2015-10-23 | | Release date: | 2016-10-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.674 Å) | | Cite: | Small Glycols Discover Cryptic Pockets on Proteins for Fragment-Based Approaches.
J.Chem.Inf.Model., 2021
|
|
