1CJD
 
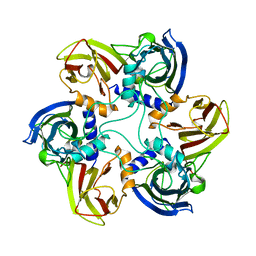 | | THE BACTERIOPHAGE PRD1 COAT PROTEIN, P3, IS STRUCTURALLY SIMILAR TO HUMAN ADENOVIRUS HEXON | | Descriptor: | PROTEIN (MAJOR CAPSID PROTEIN (P3)) | | Authors: | Benson, S.D, Bamford, J.K.H, Bamford, D.H, Burnett, R.M. | | Deposit date: | 1999-04-12 | | Release date: | 1999-09-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Viral evolution revealed by bacteriophage PRD1 and human adenovirus coat protein structures.
Cell(Cambridge,Mass.), 98, 1999
|
|
1N7U
 
 | | THE RECEPTOR-BINDING PROTEIN P2 OF BACTERIOPHAGE PRD1: CRYSTAL FORM I | | Descriptor: | ACETATE ION, Adsorption protein P2, CALCIUM ION | | Authors: | Xu, L, Benson, S.D, Butcher, S.J, Bamford, D.H, Burnett, R.M. | | Deposit date: | 2002-11-18 | | Release date: | 2003-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Receptor Binding Protein P2 of PRD1, a
Virus Targeting Antibiotic-Resistant Bacteria,
Has a Novel Fold Suggesting Multiple Functions.
Structure, 11, 2003
|
|
1N7V
 
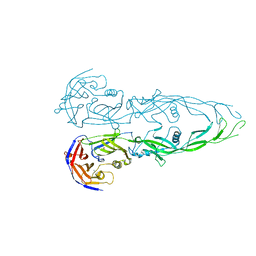 | | THE RECEPTOR-BINDING PROTEIN P2 OF BACTERIOPHAGE PRD1: CRYSTAL FORM III | | Descriptor: | ACETATE ION, Adsorption protein P2, CALCIUM ION | | Authors: | Xu, L, Benson, S.D, Butcher, S.J, Bamford, D.H, Burnett, R.M. | | Deposit date: | 2002-11-18 | | Release date: | 2003-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Receptor Binding Protein P2 of PRD1, a
Virus Targeting Antibiotic-Resistant Bacteria,
Has a Novel Fold Suggesting Multiple Functions.
Structure, 11, 2003
|
|
2W0C
 
 | | X-ray structure of the entire lipid-containing bacteriophage PM2 | | Descriptor: | CALCIUM ION, MAJOR CAPSID PROTEIN P2, PROTEIN 2, ... | | Authors: | Abrescia, N.G.A, Grimes, J.M, Kivela, H.M, Assenberg, R, Sutton, G.C, Butcher, S.J, Bamford, J.K.H, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2008-08-13 | | Release date: | 2008-09-23 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (7 Å) | | Cite: | Insights Into Virus Evolution and Membrane Biogenesis from the Structure of the Marine Lipid-Containing Bacteriophage Pm2
Mol.Cell, 31, 2008
|
|
2VVD
 
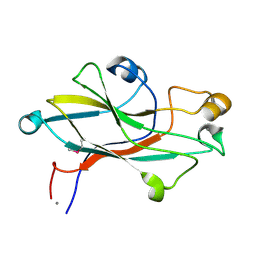 | | Crystal structure of the receptor binding domain of the spike protein P1 from bacteriophage PM2 | | Descriptor: | CALCIUM ION, SPIKE PROTEIN P1 | | Authors: | Abrescia, N.G.A, Grimes, J.M, Kivela, H.K, Assenberg, R, Sutton, G.C, Butcher, S.J, Bamford, J.K.H, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2008-06-06 | | Release date: | 2008-09-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Insights Into Virus Evolution and Membrane Biogenesis from the Structure of the Marine Lipid-Containing Bacteriophage Pm2.
Mol.Cell, 31, 2008
|
|
2VVF
 
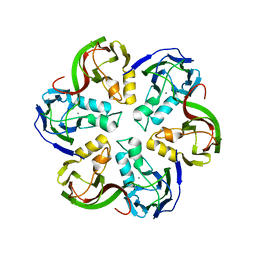 | | Crystal structure of the major capsid protein P2 from Bacteriophage PM2 | | Descriptor: | CALCIUM ION, MAJOR CAPSID PROTEIN P2 | | Authors: | Abrescia, N.G.A, Grimes, J.M, Kivela, H.K, Assenberg, R, Sutton, G.C, Butcher, S.J, Bamford, J.K.H, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2008-06-06 | | Release date: | 2008-09-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights Into Virus Evolution and Membrane Biogenesis from the Structure of the Marine Lipid-Containing Bacteriophage Pm2.
Mol.Cell, 31, 2008
|
|
1GW8
 
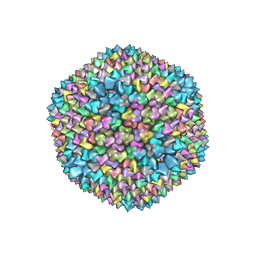 | | quasi-atomic resolution model of bacteriophage PRD1 sus607 mutant, obtained by combined cryo-EM and X-ray crystallography. | | Descriptor: | MAJOR CAPSID PROTEIN | | Authors: | San Martin, C, Huiskonen, J, Bamford, J.K.H, Butcher, S.J, Fuller, S.D, Bamford, D.H, Burnett, R.M. | | Deposit date: | 2002-03-08 | | Release date: | 2002-03-15 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (13.3 Å) | | Cite: | Minor Proteins, Mobile Arms and Membrane-Capsid Interactions in the Bacteriophage Prd1 Capsid.
Nat.Struct.Biol., 9, 2002
|
|
1GW7
 
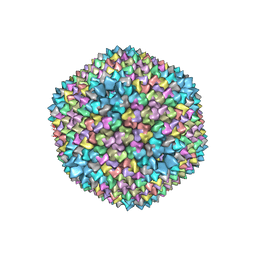 | | QUASI-ATOMIC RESOLUTION MODEL OF BACTERIOPHAGE PRD1 CAPSID, OBTAINED BY COMBINED CRYO-EM AND X-RAY CRYSTALLOGRAPHY. | | Descriptor: | MAJOR CAPSID PROTEIN | | Authors: | San Martin, C, Huiskonen, J, Bamford, J.K.H, Butcher, S.J, Fuller, S.D, Bamford, D.H, Burnett, R.M. | | Deposit date: | 2002-03-08 | | Release date: | 2002-03-13 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (13.5 Å) | | Cite: | Minor Proteins, Mobile Arms, and Membrane-Capsid Interactions in Bacteriophage Prd1 Capsid Assembly
Nat.Struct.Biol., 9, 2002
|
|
1HX6
 
 | | P3, THE MAJOR COAT PROTEIN OF THE LIPID-CONTAINING BACTERIOPHAGE PRD1. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, MAJOR CAPSID PROTEIN, ... | | Authors: | Benson, S.D, Bamford, J.K.H, Bamford, D.H, Burnett, R.M. | | Deposit date: | 2001-01-11 | | Release date: | 2001-01-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The X-ray crystal structure of P3, the major coat protein of the lipid-containing bacteriophage PRD1, at 1.65 A resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1HQN
 
 | | THE SELENOMETHIONINE DERIVATIVE OF P3, THE MAJOR COAT PROTEIN OF THE LIPID-CONTAINING BACTERIOPHAGE PRD1. | | Descriptor: | MAJOR CAPSID PROTEIN | | Authors: | Benson, S.D, Bamford, J.K.H, Bamford, D.H, Burnett, R.M. | | Deposit date: | 2000-12-18 | | Release date: | 2001-01-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The X-ray crystal structure of P3, the major coat protein of the lipid-containing bacteriophage PRD1, at 1.65 A resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
2VVE
 
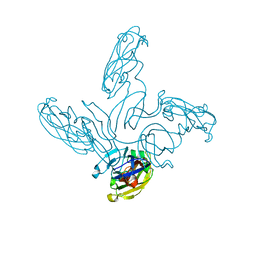 | | Crystal structure of the stem and receptor binding domain of the spike protein P1 from bacteriophage PM2 | | Descriptor: | CALCIUM ION, CHLORIDE ION, SPIKE PROTEIN P1 | | Authors: | Abrescia, N.G.A, Grimes, J.M, Kivela, H.K, Assenberg, R, Sutton, G.C, Butcher, S.J, Bamford, J.K.H, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2008-06-06 | | Release date: | 2008-09-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Insights Into Virus Evolution and Membrane Biogenesis from the Structure of the Marine Lipid-Containing Bacteriophage Pm2.
Mol.Cell, 31, 2008
|
|
1HB7
 
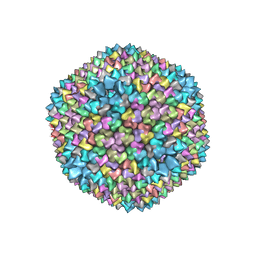 | | quasi-atomic resolution model of bacteriophage PRD1 sus1 mutant, obtained by combined cryo-EM and X-ray crystallography. | | Descriptor: | BACTERIOPHAGE PRD1 SUS1 MUTANT CAPSID | | Authors: | San Martin, C, Burnett, R.M, De Haas, F, Heinkel, R, Rutten, T, Fuller, S.D, Butcher, S.J, Bamford, D.H. | | Deposit date: | 2001-04-12 | | Release date: | 2001-12-05 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Combined Em/X-Ray Imaging Yields a Quasi-Atomic Model of the Adenovirus-Related Bacteriophage Prd1 and Shows Key Capsid and Membrane Interactions.
Structure, 9, 2001
|
|
1HB9
 
 | | quasi-atomic resolution model of bacteriophage PRD1 wild type virion, obtained by combined cryo-EM and X-ray crystallography. | | Descriptor: | BACTERIOPHAGE PRD1 | | Authors: | San Martin, C, Burnett, R.M, De Haas, F, Heinkel, R, Rutten, T, Fuller, S.D, Butcher, S.J, Bamford, D.H. | | Deposit date: | 2001-04-13 | | Release date: | 2001-12-05 | | Last modified: | 2018-01-31 | | Method: | ELECTRON MICROSCOPY (25 Å) | | Cite: | Combined Em/X-Ray Imaging Yields a Quasi-Atomic Model of the Adenovirus-Related Bacteriophage Prd1 and Shows Key Capsid and Membrane Interactions
Structure, 9, 2001
|
|
1HB5
 
 | | quasi-atomic resolution model of bacteriophage PRD1 P3-shell, obtained by combined cryo-EM and X-ray crystallography. | | Descriptor: | BACTERIOPHAGE PRD1 P3-SHELL | | Authors: | San Martin, C, Burnett, R.M, De Haas, F, Heinkel, R, Rutten, T, Fuller, S.D, Butcher, S.J, Bamford, D.H. | | Deposit date: | 2001-04-11 | | Release date: | 2001-12-05 | | Last modified: | 2018-01-31 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Combined Em/X-Ray Imaging Yields a Quasi-Atomic Model of the Adenovirus-Related Bacteriophage Prd1 and Shows Key Capsid and Membrane Interactions.
Structure, 9, 2001
|
|
2J7N
 
 | | Structure of the RNAi polymerase from Neurospora crassa | | Descriptor: | GLYCEROL, MAGNESIUM ION, RNA-DEPENDENT RNA POLYMERASE | | Authors: | Salgado, P.S, Koivunen, M.R.L, Makeyev, E.V, Bamford, D.H, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2006-10-13 | | Release date: | 2006-12-13 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structure of an Rnai Polymerase Links RNA Silencing and Transcription.
Plos Biol., 4, 2006
|
|
2VHC
 
 | | P4 PROTEIN FROM BACTERIOPHAGE PHI12 N234G mutant in complex with AMPCPP and MN | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MANGANESE (II) ION, NTPASE P4 | | Authors: | Kainov, D.E, Mancini, E.J, Telenius, J, Lisal, J, Grimes, J.M, Bamford, D.H, Stuart, D.I, Tuma, R. | | Deposit date: | 2007-11-20 | | Release date: | 2007-12-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis of Mechanochemical Coupling in a Hexameric Molecular Motor.
J.Biol.Chem., 283, 2008
|
|
2VHQ
 
 | | P4 PROTEIN FROM BACTERIOPHAGE PHI12 S252A mutant in complex with ATP AND MG | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, NTPASE P4 | | Authors: | Kainov, D.E, Mancini, E.J, Telenius, J, Lisal, J, Grimes, J.M, Bamford, D.H, Stuart, D.I, Tuma, R. | | Deposit date: | 2007-11-22 | | Release date: | 2007-12-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Basis of Mechanochemical Coupling in a Hexameric Molecular Motor.
J.Biol.Chem., 283, 2008
|
|
2VHJ
 
 | | P4 PROTEIN FROM BACTERIOPHAGE PHI12 S252A mutant in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, NTPASE P4 | | Authors: | Kainov, D.E, Mancini, E.J, Telenius, J, Lisal, J, Grimes, J.M, Bamford, D.H, Stuart, D.I, Tuma, R. | | Deposit date: | 2007-11-21 | | Release date: | 2007-12-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Mechanochemical Coupling in a Hexameric Molecular Motor.
J.Biol.Chem., 283, 2008
|
|
2VHT
 
 | | P4 PROTEIN FROM BACTERIOPHAGE PHI12 R279A mutant in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, NTPASE P4 | | Authors: | Kainov, D.E, Mancini, E.J, Telenius, J, Lisal, J, Grimes, J.M, Bamford, D.H, Stuart, D.I, Tuma, R. | | Deposit date: | 2007-11-25 | | Release date: | 2007-12-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis of Mechanochemical Coupling in a Hexameric Molecular Motor.
J.Biol.Chem., 283, 2008
|
|
2VHU
 
 | | P4 PROTEIN FROM BACTERIOPHAGE PHI12 K241C mutant in complex with ADP and MgCl | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NTPASE P4 | | Authors: | Kainov, D.E, Mancini, E.J, Telenius, J, Lisal, J, Grimes, J.M, Bamford, D.H, Stuart, D.I, Tuma, R. | | Deposit date: | 2007-11-25 | | Release date: | 2007-12-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Basis of Mechanochemical Coupling in a Hexameric Molecular Motor.
J.Biol.Chem., 283, 2008
|
|
2YI8
 
 | | Structure of the RNA polymerase VP1 from Infectious Pancreatic Necrosis Virus | | Descriptor: | CHLORIDE ION, POTASSIUM ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Graham, S.C, Sarin, L.P, Bahar, M.W, Myers, R.A, Stuart, D.I, Bamford, D.H, Grimes, J.M. | | Deposit date: | 2011-05-11 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The N-Terminus of the RNA Polymerase from Infectious Pancreatic Necrosis Virus is the Determinant of Genome Attachment.
Plos Pathog., 7, 2011
|
|
2YI9
 
 | | Structure of the RNA polymerase VP1 from Infectious Pancreatic Necrosis Virus in complex with magnesium | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Graham, S.C, Sarin, L.P, Bahar, M.W, Myers, R.A, Stuart, D.I, Bamford, D.H, Grimes, J.M. | | Deposit date: | 2011-05-11 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The N-Terminus of the RNA Polymerase from Infectious Pancreatic Necrosis Virus is the Determinant of Genome Attachment.
Plos Pathog., 7, 2011
|
|
2YIA
 
 | | Structure of the RNA polymerase VP1 from Infectious Pancreatic Necrosis Virus | | Descriptor: | POTASSIUM ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Graham, S.C, Sarin, L.P, Bahar, M.W, Myers, R.A, Stuart, D.I, Bamford, D.H, Grimes, J.M. | | Deposit date: | 2011-05-11 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | The N-Terminus of the RNA Polymerase from Infectious Pancreatic Necrosis Virus is the Determinant of Genome Attachment.
Plos Pathog., 7, 2011
|
|
2YIB
 
 | | Structure of the RNA polymerase VP1 from Infectious Pancreatic Necrosis Virus | | Descriptor: | RNA-DIRECTED RNA POLYMERASE | | Authors: | Graham, S.C, Sarin, L.P, Bahar, M.W, Myers, R.A, Stuart, D.I, Bamford, D.H, Grimes, J.M. | | Deposit date: | 2011-05-11 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The N-Terminus of the RNA Polymerase from Infectious Pancreatic Necrosis Virus is the Determinant of Genome Attachment.
Plos Pathog., 7, 2011
|
|
2J7O
 
 | | STRUCTURE OF THE RNAI POLYMERASE FROM NEUROSPORA CRASSA | | Descriptor: | MAGNESIUM ION, RNA DEPENDENT RNA POLYMERASE | | Authors: | Salgado, P.S, Koivunen, M.R.L, Makeyev, E.V, Bamford, D.H, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2006-10-13 | | Release date: | 2006-12-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The Structure of an Rnai Polymerase Links RNA Silencing and Transcription.
Plos Biol., 4, 2006
|
|
