1PRR
 
 | | NMR-DERIVED THREE-DIMENSIONAL SOLUTION STRUCTURE OF PROTEIN S COMPLEXED WITH CALCIUM | | Descriptor: | CALCIUM ION, DEVELOPMENT-SPECIFIC PROTEIN S | | Authors: | Bagby, S, Harvey, T.S, Eagle, S.G, Inouye, S, Ikura, M. | | Deposit date: | 1994-03-25 | | Release date: | 1994-08-31 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR-derived three-dimensional solution structure of protein S complexed with calcium.
Structure, 2, 1994
|
|
1PRS
 
 | | NMR-DERIVED THREE-DIMENSIONAL SOLUTION STRUCTURE OF PROTEIN S COMPLEXED WITH CALCIUM | | Descriptor: | CALCIUM ION, DEVELOPMENT-SPECIFIC PROTEIN S | | Authors: | Bagby, S, Harvey, T.S, Eagle, S.G, Inouye, S, Ikura, M. | | Deposit date: | 1994-03-25 | | Release date: | 1994-08-31 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR-derived three-dimensional solution structure of protein S complexed with calcium.
Structure, 2, 1994
|
|
1PLA
 
 | | HIGH-RESOLUTION SOLUTION STRUCTURE OF REDUCED PARSLEY PLASTOCYANIN | | Descriptor: | COPPER (II) ION, PLASTOCYANIN | | Authors: | Bagby, S, Driscoll, P.C, Harvey, T.S, Hill, H.A.O. | | Deposit date: | 1994-05-20 | | Release date: | 1994-08-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of reduced parsley plastocyanin.
Biochemistry, 33, 1994
|
|
1PLB
 
 | | HIGH-RESOLUTION SOLUTION STRUCTURE OF REDUCED PARSLEY PLASTOCYANIN | | Descriptor: | COPPER (II) ION, PLASTOCYANIN | | Authors: | Bagby, S, Driscoll, P.C, Harvey, T.S, Hill, H.A.O. | | Deposit date: | 1994-05-20 | | Release date: | 1994-08-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of reduced parsley plastocyanin.
Biochemistry, 33, 1994
|
|
1TFB
 
 | |
2L5T
 
 | |
2WY7
 
 | | Staphylococcus aureus complement subversion protein Sbi-IV in complex with complement fragment C3d revealing an alternative binding mode | | Descriptor: | COMPLEMENT C3D FRAGMENT, GLYCEROL, IGG-BINDING PROTEIN | | Authors: | Clark, E.A, Crennell, S, Upadhyay, A, Mackay, J.D, Bagby, S, van den Elsen, J.M. | | Deposit date: | 2009-11-13 | | Release date: | 2010-12-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Structural Basis for Staphylococcal Complement Subversion: X-Ray Structure of the Complement- Binding Domain of Staphylococcus Aureus Protein Sbi in Complex with Ligand C3D.
Mol.Immunol., 48, 2011
|
|
2WY8
 
 | | Staphylococcus aureus complement subversion protein Sbi-IV in complex with complement fragment C3d | | Descriptor: | COMPLEMENT C3D FRAGMENT, GLYCEROL, IGG-BINDING PROTEIN | | Authors: | Clark, E.A, Crennell, S, Upadhyay, A, Mackay, J.D, Bagby, S, van den Elsen, J.M. | | Deposit date: | 2009-11-13 | | Release date: | 2010-12-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Structural Basis for Staphylococcal Complement Subversion: X-Ray Structure of the Complement- Binding Domain of Staphylococcus Aureus Protein Sbi in Complex with Ligand C3D.
Mol.Immunol., 48, 2011
|
|
6FB4
 
 | | human KIBRA C2 domain mutant C771A | | Descriptor: | GLYCEROL, PHOSPHATE ION, Protein KIBRA | | Authors: | Crennell, S.J, Posner, M.G, Bagby, S. | | Deposit date: | 2017-12-18 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.415631 Å) | | Cite: | Distinctive phosphoinositide- and Ca2+-binding properties of normal and cognitive performance-linked variant forms of KIBRA C2 domain.
J. Biol. Chem., 293, 2018
|
|
6FJC
 
 | | Human KIBRA C2 domain mutant C771A in complex with phosphatidylinositol 3,4,5-trisphosphate | | Descriptor: | (2R)-3-{[(S)-{[(2S,3R,5S,6S)-2,6-DIHYDROXY-3,4,5-TRIS(PHOSPHONOOXY)CYCLOHEXYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-(1-HYDROXY BUTOXY)PROPYL BUTYRATE, GLYCEROL, Protein KIBRA, ... | | Authors: | Crennell, S.J, Posner, M.G, Bagby, S. | | Deposit date: | 2018-01-22 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Distinctive phosphoinositide- and Ca2+-binding properties of normal and cognitive performance-linked variant forms of KIBRA C2 domain.
J. Biol. Chem., 293, 2018
|
|
6FJD
 
 | | Human KIBRA C2 domain mutant C771A in complex with phosphatidylinositol 4,5-bisphosphate | | Descriptor: | (2R)-3-{[(R)-HYDROXY{[(1R,2R,3S,4R,5R,6S)-2,3,6-TRIHYDROXY-4,5-BIS(PHOSPHONOOXY)CYCLOHEXYL]OXY}PHOSPHORYL]OXY}PROPANE-1 ,2-DIYL DIBUTANOATE, GLYCEROL, Protein KIBRA, ... | | Authors: | Crennell, S.J, Posner, M.G, Bagby, S. | | Deposit date: | 2018-01-22 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.898 Å) | | Cite: | Distinctive phosphoinositide- and Ca2+-binding properties of normal and cognitive performance-linked variant forms of KIBRA C2 domain.
J. Biol. Chem., 293, 2018
|
|
6FD0
 
 | | Human KIBRA C2 domain mutant M734I S735A | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Protein KIBRA | | Authors: | Crennell, S.J, Posner, M.G, Bagby, S. | | Deposit date: | 2017-12-21 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.64215851 Å) | | Cite: | Distinctive phosphoinositide- and Ca2+-binding properties of normal and cognitive performance-linked variant forms of KIBRA C2 domain.
J. Biol. Chem., 293, 2018
|
|
1R9K
 
 | | Representative solution structure of the catalytic domain of SopE2 | | Descriptor: | TypeIII-secreted protein effector: invasion-associated protein | | Authors: | Williams, C, Galyov, E.E, Bagby, S. | | Deposit date: | 2003-10-30 | | Release date: | 2004-09-28 | | Last modified: | 2018-01-24 | | Method: | SOLUTION NMR | | Cite: | Solution Structure, Backbone Dynamics, and Interaction with Cdc42 of Salmonella Guanine Nucleotide Exchange Factor SopE2(,).
Biochemistry, 43, 2004
|
|
2JVH
 
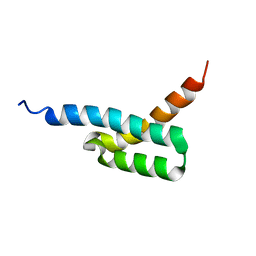 | | Structure of C3-binding domain 4 of S. aureus protein Sbi | | Descriptor: | IgG-binding protein SBI | | Authors: | Upadhyay, A, Burman, J, Clark, E.A, van den Elsen, J.M.H, Bagby, S. | | Deposit date: | 2007-09-20 | | Release date: | 2008-06-10 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of the C3 binding region of Staphylococcus aureus immune subversion protein Sbi.
J.Biol.Chem., 283, 2008
|
|
2JVG
 
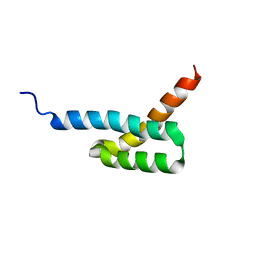 | | Structure of C3-binding domain 4 of Staphylococcus aureus protein Sbi | | Descriptor: | IgG-binding protein SBI | | Authors: | Upadhyay, A, Burman, J, Clark, E.A, van den Elsen, J.M.H, Bagby, S. | | Deposit date: | 2007-09-20 | | Release date: | 2008-06-10 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of the C3 binding region of Staphylococcus aureus immune subversion protein Sbi.
J.Biol.Chem., 283, 2008
|
|
2JOK
 
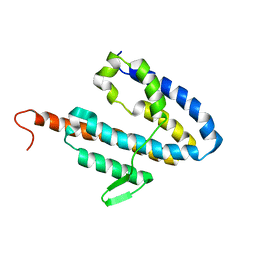 | | NMR structure of the catalytic domain of guanine nucleotide exchange factor BopE from Burkholderia pseudomallei | | Descriptor: | Putative G-nucleotide exchange factor | | Authors: | Wu, H, Upadhyay, A, Williams, C, Galyov, E.E, van den Elsen, J.M.H, Bagby, S. | | Deposit date: | 2007-03-14 | | Release date: | 2007-09-18 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | The guanine-nucleotide-exchange factor BopE from Burkholderia pseudomallei adopts a compact version of the Salmonella SopE/SopE2 fold and undergoes a closed-to-open conformational change upon interaction with Cdc42
Biochem.J., 411, 2008
|
|
2JOL
 
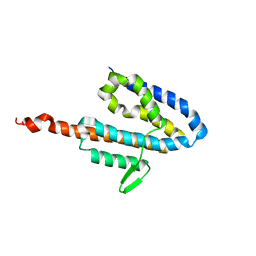 | | Average NMR structure of the catalytic domain of guanine nucleotide exchange factor BopE from Burkholderia pseudomallei | | Descriptor: | Putative G-nucleotide exchange factor | | Authors: | Wu, H, Upadhyay, A, Williams, C, Galyov, E.E, van den Elsen, J.M.H, Bagby, S. | | Deposit date: | 2007-03-14 | | Release date: | 2007-03-27 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | The guanine-nucleotide-exchange factor BopE from Burkholderia pseudomallei adopts a compact version of the Salmonella SopE/SopE2 fold and undergoes a closed-to-open conformational change upon interaction with Cdc42
Biochem.J., 411, 2008
|
|
2L4J
 
 | | Yap ww2 | | Descriptor: | Yes-associated protein 2 (YAP2) | | Authors: | Webb, C, Upadhyay, A, Furutani-Seiki, M, Bagby, S. | | Deposit date: | 2010-10-07 | | Release date: | 2010-11-03 | | Last modified: | 2020-02-05 | | Method: | SOLUTION NMR | | Cite: | Structural Features and Ligand Binding Properties of Tandem WW Domains from YAP and TAZ, Nuclear Effectors of the Hippo Pathway.
Biochemistry, 50, 2011
|
|
1R6E
 
 | |
1TBA
 
 | | SOLUTION STRUCTURE OF A TBP-TAFII230 COMPLEX: PROTEIN MIMICRY OF THE MINOR GROOVE SURFACE OF THE TATA BOX UNWOUND BY TBP, NMR, 25 STRUCTURES | | Descriptor: | TRANSCRIPTION INITIATION FACTOR IID 230K CHAIN, TRANSCRIPTION INITIATION FACTOR TFIID | | Authors: | Liu, D, Ishima, R, Tong, K.I, Bagby, S, Kokubo, T, Muhandiram, D.R, Kay, L.E, Nakatani, Y, Ikura, M. | | Deposit date: | 1998-08-16 | | Release date: | 1999-08-16 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a TBP-TAF(II)230 complex: protein mimicry of the minor groove surface of the TATA box unwound by TBP.
Cell(Cambridge,Mass.), 94, 1998
|
|
3R07
 
 | |
1C9B
 
 | |
1NPS
 
 | | CRYSTAL STRUCTURE OF N-TERMINAL DOMAIN OF PROTEIN S | | Descriptor: | CALCIUM ION, DEVELOPMENT-SPECIFIC PROTEIN S | | Authors: | Wenk, M, Baumgartner, R, Mayer, E.M, Huber, R, Holak, T.A, Jaenicke, R. | | Deposit date: | 1999-02-01 | | Release date: | 2000-02-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The domains of protein S from Myxococcus xanthus: structure, stability and interactions.
J.Mol.Biol., 286, 1999
|
|
1SUH
 
 | | AMINO-TERMINAL DOMAIN OF EPITHELIAL CADHERIN IN THE CALCIUM BOUND STATE, NMR, 20 STRUCTURES | | Descriptor: | EPITHELIAL CADHERIN | | Authors: | Overduin, M, Tong, K.I, Kay, C.M, Ikura, M. | | Deposit date: | 1996-01-30 | | Release date: | 1996-07-11 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | 1H, 15N and 13C resonance assignments and monomeric structure of the amino-terminal extracellular domain of epithelial cadherin.
J.Biomol.NMR, 7, 1996
|
|
