458D
 
 | |
459D
 
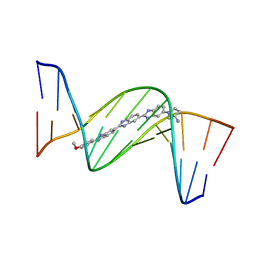 | | DNA MINOR-GROOVE RECOGNITION OF A TRIS-BENZIMIDAZOLE DRUG | | Descriptor: | 2''-(4-METHOXYPHENYL)-5-(3-AMINO-1-PYRROLIDINYL)-2,5',2',5''-TRI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*(CBR)P*AP*TP*AP*TP*TP*TP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*AP*AP*AP*TP*AP*TP*GP*CP*G)-3') | | Authors: | Aymami, J, Nunn, C.M, Neidle, S. | | Deposit date: | 1999-03-10 | | Release date: | 1999-06-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA minor groove recognition of a non-self-complementary AT-rich sequence by a tris-benzimidazole ligand.
Nucleic Acids Res., 27, 1999
|
|
121D
 
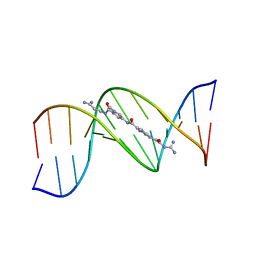 | | MOLECULAR STRUCTURE OF THE A-TRACT DNA DODECAMER D(CGCAAATTTGCG) COMPLEXED WITH THE MINOR GROOVE BINDING DRUG NETROPSIN | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*AP*AP*TP*TP*TP*GP*CP*G)-3'), NETROPSIN | | Authors: | Tabernero, L, Verdaguer, N, Coll, M, Fita, I, Van Der Marel, G.A, Van Boom, J.H, Rich, A, Aymami, J. | | Deposit date: | 1993-04-14 | | Release date: | 1994-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular structure of the A-tract DNA dodecamer d(CGCAAATTTGCG) complexed with the minor groove binding drug netropsin.
Biochemistry, 32, 1993
|
|
1K9G
 
 | | Crystal Structure of the Complex of Cryptolepine-d(CCTAGG)2 | | Descriptor: | 5'-D(*CP*CP*TP*AP*GP*G)-3', 5-METHYL-5H-INDOLO[3,2-B]QUINOLINE | | Authors: | Lisgarten, J.N, Coll, M, Portugal, J, Wright, C.W, Aymami, J. | | Deposit date: | 2001-10-29 | | Release date: | 2001-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The antimalarial and cytotoxic drug cryptolepine intercalates into DNA at cytosine-cytosine sites.
Nat.Struct.Biol., 9, 2002
|
|
2ET0
 
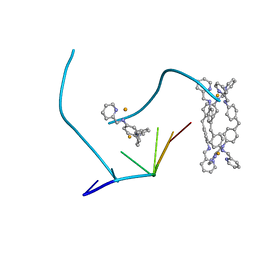 | | The structure of a three-way DNA junction in complex with a metallo-supramolecular helicate reveals a new target for drugs | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*G)-3', FE (II) ION, N-[(1E)-PYRIDIN-2-YLMETHYLENE]-N-[4-(4-{[(1E)-PYRIDIN-2-YLMETHYLENE]AMINO}BENZYL)PHENYL]AMINE | | Authors: | Oleksi, A, Blanco, A.G, Boer, R, Uson, I, Aymami, J, Coll, M. | | Deposit date: | 2005-10-27 | | Release date: | 2006-03-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular Recognition of a Three-Way DNA Junction by a Metallosupramolecular Helicate
ANGEW.CHEM.INT.ED.ENGL., 45, 2006
|
|
264D
 
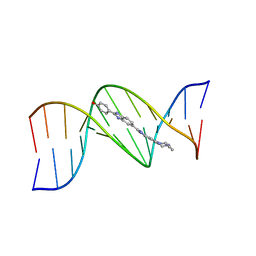 | | THREE-DIMENSIONAL CRYSTAL STRUCTURE OF THE A-TRACT DNA DODECAMER D(CGCAAATTTGCG) COMPLEXED WITH THE MINOR-GROOVE-BINDING DRUG HOECHST 33258 | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*AP*AP*AP*TP*TP*TP*GP*CP*G)-3') | | Authors: | Vega, M.C, Garcia-Saez, I, Aymami, J, Eritja, R, Van Der Marel, G.A, Van Boom, J.H, Rich, A, Coll, M. | | Deposit date: | 1994-09-22 | | Release date: | 1995-03-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Three-dimensional crystal structure of the A-tract DNA dodecamer d(CGCAAATTTGCG) complexed with the minor-groove-binding drug Hoechst 33258.
Eur.J.Biochem., 222, 1994
|
|
2D47
 
 | | MOLECULAR STRUCTURE OF A COMPLETE TURN OF A-DNA | | Descriptor: | DNA (5'-D(*CP*CP*CP*CP*CP*GP*CP*GP*GP*GP*GP*G)-3'), SPERMINE | | Authors: | Verdaguer, N, Aymami, J, Fernandez-Forner, D, Fita, I, Coll, M, Huynh-Dinh, T, Igolen, J, Subirana, J.A. | | Deposit date: | 1991-10-02 | | Release date: | 1991-10-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular structure of a complete turn of A-DNA.
J.Mol.Biol., 221, 1991
|
|
1VTJ
 
 | | MOLECULAR STRUCTURE OF THE NETROPSIN-D(CGCGATATCGCG) COMPLEX: DNA CONFORMATION IN AN ALTERNATING AT SEGMENT; CONFORMATION 1 | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*TP*AP*TP*CP*GP*CP*G)-3'), NETROPSIN | | Authors: | Coll, M, Aymami, J, Van Der Marel, G.A, Van Boom, J.H, Rich, A, Wang, A.H.-J. | | Deposit date: | 1999-09-14 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Structure of the Netropsin-d(CGCGATATCGCG) Complex: DNA Conformation in an Alternating AT Segment
Biochemistry, 28, 1989
|
|
4MNB
 
 | | Crystal Structure of a complex between the marine anticancer drug Variolin B and DNA | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*G)-3', 9-amino-5-(2-aminopyrimidin-4-yl)pyrido[3',2':4,5]pyrrolo[1,2-c]pyrimidin-4-ol, COBALT (II) ION, ... | | Authors: | Canals, A, Arribas-Bosacoma, R, Alvarez, M, Albericio, F, Aymami, J, Coll, M. | | Deposit date: | 2013-09-10 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Intercalative DNA binding of the marine anticancer drug variolin B.
Sci Rep, 7, 2017
|
|
1Z3F
 
 | | Structure of ellipticine in complex with a 6-bp DNA | | Descriptor: | 5'-D(*CP*GP*AP*TP*CP*G)-3', COBALT (II) ION, ELLIPTICINE | | Authors: | Canals, A, Purciolas, M, Aymami, J, Coll, M. | | Deposit date: | 2005-03-12 | | Release date: | 2005-07-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The anticancer agent ellipticine unwinds DNA by intercalative binding in an orientation parallel to base pairs.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1DNE
 
 | | MOLECULAR STRUCTURE OF THE NETROPSIN-D(CGCGATATCGCG) COMPLEX: DNA CONFORMATION IN AN ALTERNATING AT SEGMENT; CONFORMATION 2 | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*TP*AP*TP*CP*GP*CP*G)-3'), NETROPSIN | | Authors: | Coll, M, Aymami, J, Van Der Marel, G.A, Van Boom, J.H, Rich, A, Wang, A.H.-J. | | Deposit date: | 1988-09-14 | | Release date: | 1989-01-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular structure of the netropsin-d(CGCGATATCGCG) complex: DNA conformation in an alternating AT segment.
Biochemistry, 28, 1989
|
|
1DNF
 
 | | EFFECTS OF 5-FLUOROURACIL/GUANINE WOBBLE BASE PAIRS IN Z-DNA. MOLECULAR AND CRYSTAL STRUCTURE OF D(CGCGFG) | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*(UFP)P*G)-3'), MAGNESIUM ION | | Authors: | Coll, M, Saal, D, Frederick, C.A, Aymami, J, Rich, A, Wang, A.H.-J. | | Deposit date: | 1988-12-12 | | Release date: | 1990-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Effects of 5-fluorouracil/guanine wobble base pairs in Z-DNA: molecular and crystal structure of d(CGCGFG).
Nucleic Acids Res., 17, 1989
|
|
1G3X
 
 | | INTERCALATION OF AN 9ACRIDINE-PEPTIDE DRUG IN A DNA DODECAMER | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', MAGNESIUM ION, N(ALPHA)-(9-ACRIDINOYL)-TETRAARGININE-AMIDE | | Authors: | Malinina, L, Soler-Lopez, M, Aymami, J, Subirana, J.A. | | Deposit date: | 2000-10-25 | | Release date: | 2002-08-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Intercalation of an acridine-peptide drug in an AA/TT base step in the crystal structure of [d(CGCGAATTCGCG)](2) with six duplexes and seven Mg(2+) ions in the asymmetric unit.
Biochemistry, 41, 2002
|
|
1NDN
 
 | | MOLECULAR STRUCTURE OF NICKED DNA. MODEL T4 | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*AP*AP*CP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*GP*TP*T)-3'), DNA (5'-D(*TP*TP*CP*GP*CP*G)-3') | | Authors: | Aymani, J, Coll, M, Van Der Marel, G.A, Van Boom, J.H, Wang, A.H.-J, Rich, A. | | Deposit date: | 1992-01-15 | | Release date: | 1992-07-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular structure of nicked DNA: a substrate for DNA repair enzymes.
Proc.Natl.Acad.Sci.USA, 87, 1990
|
|
1VTE
 
 | | MOLECULAR STRUCTURE OF NICKED DNA. MODEL A4 | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*AP*AP*CP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*GP*TP*T)-3'), DNA (5'-D(*TP*TP*CP*GP*CP*G)-3') | | Authors: | Aymani, J, Coll, M, Van Der Marel, G.A, Van Boom, J.H, Wang, A.H.-J, Rich, A. | | Deposit date: | 1990-05-21 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular structure of nicked DNA: a substrate for DNA repair enzymes.
Proc. Natl. Acad. Sci. U.S.A., 87, 1990
|
|
4X08
 
 | | Structure of H128N/ECP mutant in complex with sulphate anions at 1.34 Angstroms. | | Descriptor: | Eosinophil cationic protein, SULFATE ION | | Authors: | Blanco, J.A, Garcia, J.M, Salazar, V.A, Sanchez, D, Moussauoi, M, Boix, E. | | Deposit date: | 2014-11-21 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structure of H128N/ECP mutant in complex with sulphate anions at 1.34 Angstroms.
To Be Published
|
|
5K1I
 
 | | PDE4 crystal structure in complex with small molecule inhibitor | | Descriptor: | 4-[(5-acetyl-2-ethyl-3-oxo-6-phenyl-2,3-dihydropyridazin-4-yl)amino]benzoic acid, MAGNESIUM ION, ZINC ION, ... | | Authors: | Segarra, V, Hernandez, B, Ferrer-Miralles, N, Korndoerfer, I, Aymami, J. | | Deposit date: | 2016-05-18 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Biphenyl Pyridazinone Derivatives as Inhaled PDE4 Inhibitors: Structural Biology and Structure-Activity Relationships.
J. Med. Chem., 59, 2016
|
|
5K32
 
 | | PDE4D crystal structure in complex with small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-[(3-methoxyphenyl)amino]-2-phenyl-7,8-dihydro-1,6-naphthyridin-5(6H)-one, MAGNESIUM ION, ... | | Authors: | Segarra, V, Hernandez, B, Roberts, R, Gracia, J, Soler, M, Bonin, I, Aymami, J. | | Deposit date: | 2016-05-19 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | 4-Amino-7,8-dihydro-1,6-naphthyridin-5(6 H)-ones as Inhaled Phosphodiesterase Type 4 (PDE4) Inhibitors: Structural Biology and Structure-Activity Relationships.
J. Med. Chem., 61, 2018
|
|
467D
 
 | |
8F2Y
 
 | |
4OXF
 
 | |
3FX8
 
 | | Distinct recognition of three-way DNA junctions by a thioester variant of a metallo-supramolecular cylinder ('helicate') | | Descriptor: | (5'-D(*CP*GP*TP*AP*CP*G)-3', 4,4'-sulfanediylbis{N-[(1E)-pyridin-2-ylmethylidene]aniline}, FE (II) ION | | Authors: | Boer, D.R, Uson, I, Coll, M. | | Deposit date: | 2009-01-20 | | Release date: | 2010-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Self-Assembly of Functionalizable Two-Component 3D DNA Arrays through the Induced Formation of DNA Three-Way-Junction Branch Points by Supramolecular Cylinders.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
263D
 
 | | ISOHELICITY AND PHASING IN DRUG-DNA SEQUENCE RECOGNITION: CRYSTAL STRUCTURE OF A TRIS(BENZIMIDAZOLE)-OLIGONUCLEOTIDE COMPLEX | | Descriptor: | 2''-(4-METHOXYPHENYL)-5-(3-AMINO-1-PYRROLIDINYL)-2,5',2',5''-TRI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*AP*AP*AP*TP*TP*TP*GP*CP*G)-3') | | Authors: | Clark, G.R, Gray, E.J, Neidle, S, Li, Y.-H, Leupin, W. | | Deposit date: | 1996-09-27 | | Release date: | 1996-10-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Isohelicity and phasing in drug--DNA sequence recognition: crystal structure of a tris(benzimidazole)--oligonucleotide complex.
Biochemistry, 35, 1996
|
|
