4U3J
 
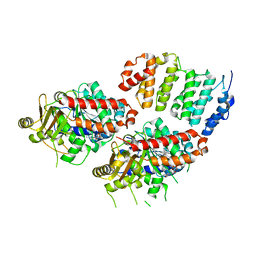 | | TOG2:alpha/beta-tubulin complex | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein STU2, ... | | Authors: | Ayaz, P, Rice, L.M. | | Deposit date: | 2014-07-22 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | A tethered delivery mechanism explains the catalytic action of a microtubule polymerase.
Elife, 3, 2014
|
|
4FFB
 
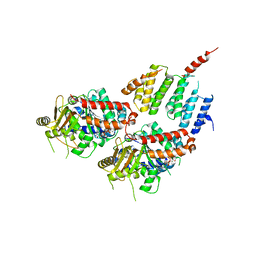 | | A TOG:alpha/beta-tubulin Complex Structure Reveals Conformation-Based Mechanisms For a Microtubule Polymerase | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein STU2, ... | | Authors: | Ayaz, P, Ye, X, Huddleston, P, Brautigam, C.A, Rice, L.M. | | Deposit date: | 2012-05-31 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.882 Å) | | Cite: | A TOG: alpha beta-tubulin complex structure reveals conformation-based mechanisms for a microtubule polymerase.
Science, 337, 2012
|
|
5IEX
 
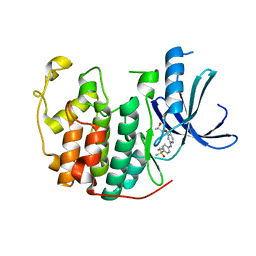 | | Crystal structure of (R,S)-S-{4-[(5-Bromo-4-{[(2R,3R)-2-hydroxy-1-methylpropyl]oxy}- pyrimidin-2-yl)amino]phenyl}-S-cyclopropylsulfoximide bound to CDK2 | | Descriptor: | (2R,3R)-3-[(5-bromo-2-{[4-(S-cyclopropylsulfonimidoyl)phenyl]amino}pyrimidin-4-yl)oxy]butan-2-ol, Cyclin-dependent kinase 2 | | Authors: | Ayaz, P, Andres, D, Kwiatkowski, D.A, Kolbe, C, Lienau, P, Siemeister, G, Luecking, U, Stegmann, C.M. | | Deposit date: | 2016-02-25 | | Release date: | 2016-04-27 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Conformational Adaption May Explain the Slow Dissociation Kinetics of Roniciclib (BAY 1000394), a Type I CDK Inhibitor with Kinetic Selectivity for CDK2 and CDK9.
Acs Chem.Biol., 11, 2016
|
|
5IF1
 
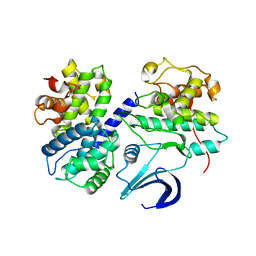 | | Crystal structure apo CDK2/cyclin A | | Descriptor: | Cyclin-A2, Cyclin-dependent kinase 2 | | Authors: | Ayaz, P, Andres, D, Kwiatkowski, D.A, Kolbe, C, Lienau, P, Siemeister, G, Luecking, U, Stegmann, C.M. | | Deposit date: | 2016-02-25 | | Release date: | 2016-04-27 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Conformational Adaption May Explain the Slow Dissociation Kinetics of Roniciclib (BAY 1000394), a Type I CDK Inhibitor with Kinetic Selectivity for CDK2 and CDK9.
Acs Chem.Biol., 11, 2016
|
|
5IEV
 
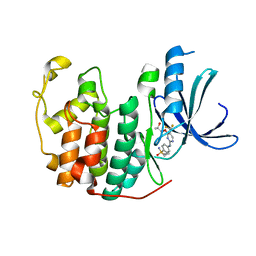 | | Crystal structure of BAY 1000394 (Roniciclib) bound to CDK2 | | Descriptor: | Cyclin-dependent kinase 2, Roniciclib | | Authors: | Ayaz, P, Andres, D, Kwiatkowski, D.A, Kolbe, C, Lienau, P, Siemeister, G, Luecking, U, Stegmann, C.M. | | Deposit date: | 2016-02-25 | | Release date: | 2016-04-27 | | Last modified: | 2022-10-26 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Conformational Adaption May Explain the Slow Dissociation Kinetics of Roniciclib (BAY 1000394), a Type I CDK Inhibitor with Kinetic Selectivity for CDK2 and CDK9.
Acs Chem.Biol., 11, 2016
|
|
5IEY
 
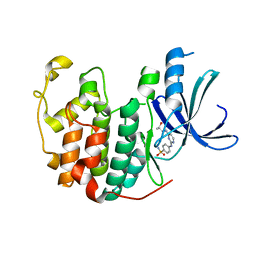 | | Crystal structure of a CDK inhibitor bound to CDK2 | | Descriptor: | 4-[(4-{[(2R,3R)-3-hydroxybutan-2-yl]amino}pyrimidin-2-yl)amino]benzene-1-sulfonamide, Cyclin-dependent kinase 2 | | Authors: | Ayaz, P, Andres, D, Kwiatkowski, D.A, Kolbe, C, Lienau, P, Siemeister, G, Luecking, U, Stegmann, C.M. | | Deposit date: | 2016-02-25 | | Release date: | 2016-04-27 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Conformational Adaption May Explain the Slow Dissociation Kinetics of Roniciclib (BAY 1000394), a Type I CDK Inhibitor with Kinetic Selectivity for CDK2 and CDK9.
Acs Chem.Biol., 11, 2016
|
|
6OP9
 
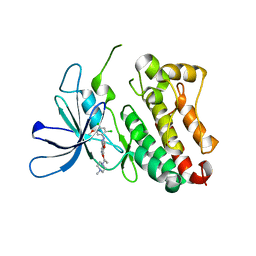 | | HER3 pseudokinase domain bound to bosutinib | | Descriptor: | 4-[(2,4-dichloro-5-methoxyphenyl)amino]-6-methoxy-7-[3-(4-methylpiperazin-1-yl)propoxy]quinoline-3-carbonitrile, Receptor tyrosine-protein kinase erbB-3 | | Authors: | Littlefield, P, Agnew, C, Jura, N. | | Deposit date: | 2019-04-24 | | Release date: | 2019-08-21 | | Last modified: | 2022-02-16 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Targetable HER3 functions driving tumorigenic signaling in HER2-amplified cancers.
Cell Rep, 38, 2022
|
|
6JXT
 
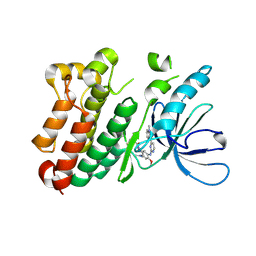 | |
6JX0
 
 | |
6JX4
 
 | |
6JWL
 
 | | Crystal structure of EGFR 696-1022 L858R in complex with AZD9291 | | Descriptor: | Epidermal growth factor receptor, N-(2-{[2-(dimethylamino)ethyl](methyl)amino}-4-methoxy-5-{[4-(1-methyl-1H-indol-3-yl)pyrimidin-2-yl]amino}phenyl)prop-2-enamide | | Authors: | Yun, C.H, Zhu, S.J, Yan, X.E. | | Deposit date: | 2019-04-21 | | Release date: | 2020-04-22 | | Last modified: | 2020-11-04 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Structural Basis of AZD9291 Selectivity for EGFR T790M.
J.Med.Chem., 63, 2020
|
|
8STG
 
 | |
8SWE
 
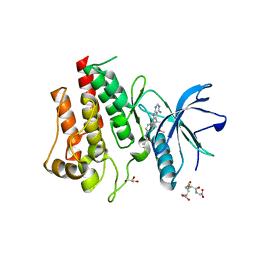 | | FGFR2 Kinase Domain Bound to Reversible Inhibitor Cmpd 3 | | Descriptor: | Fibroblast growth factor receptor 2, GLUTATHIONE, GLYCEROL, ... | | Authors: | Valverde, R, Foster, L. | | Deposit date: | 2023-05-18 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Discovery of lirafugratinib (RLY-4008), a highly selective irreversible small-molecule inhibitor of FGFR2.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8U1F
 
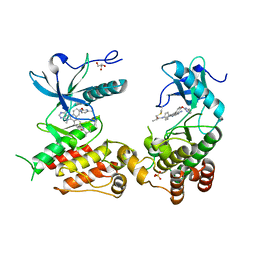 | | FGFR2 Kinase Domain Bound to Irreversible Inhibitor Cmpd 10 | | Descriptor: | Fibroblast growth factor receptor 2, GLYCEROL, N-[4-(4-amino-7-methyl-5-{4-[(4-methylpyrimidin-2-yl)oxy]phenyl}-7H-pyrrolo[2,3-d]pyrimidin-6-yl)phenyl]-2-methylpropanamide, ... | | Authors: | Valverde, R, Foster, L. | | Deposit date: | 2023-08-31 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Discovery of lirafugratinib (RLY-4008), a highly selective irreversible small-molecule inhibitor of FGFR2.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6UNQ
 
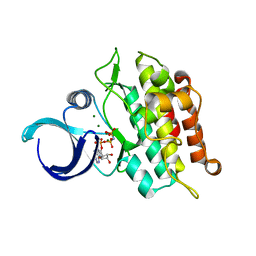 | | Kinase domain of ALK2-K493A with AMPPNP | | Descriptor: | Activin receptor type-1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Agnew, C, Jura, N. | | Deposit date: | 2019-10-13 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for ALK2/BMPR2 receptor complex signaling through kinase domain oligomerization.
Nat Commun, 12, 2021
|
|
6UNR
 
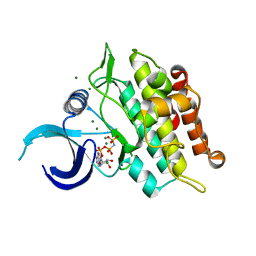 | | Kinase domain of ALK2-K492A/K493A with AMPPNP | | Descriptor: | Activin receptor type-1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Agnew, C, Jura, N. | | Deposit date: | 2019-10-13 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for ALK2/BMPR2 receptor complex signaling through kinase domain oligomerization.
Nat Commun, 12, 2021
|
|
6UNP
 
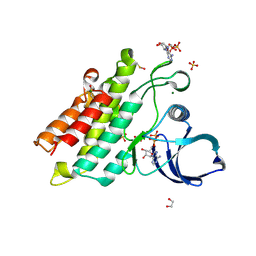 | | Crystal structure of the kinase domain of BMPR2-D485G | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, Bone morphogenetic protein receptor type-2, ... | | Authors: | Agnew, C, Jura, N. | | Deposit date: | 2019-10-13 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for ALK2/BMPR2 receptor complex signaling through kinase domain oligomerization.
Nat Commun, 12, 2021
|
|
6UNS
 
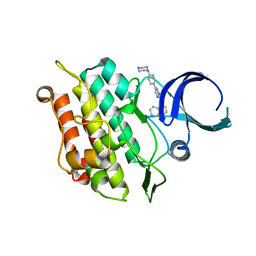 | | Kinase domain of ALK2-K492A/K493A with LDN-193189 | | Descriptor: | 4-[6-(4-piperazin-1-ylphenyl)pyrazolo[1,5-a]pyrimidin-3-yl]quinoline, Activin receptor type-1 | | Authors: | Agnew, C, Jura, N. | | Deposit date: | 2019-10-13 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for ALK2/BMPR2 receptor complex signaling through kinase domain oligomerization.
Nat Commun, 12, 2021
|
|
