5YK9
 
 | |
5Z43
 
 | | Crystal structure of prenyltransferase AmbP1 apo structure | | Descriptor: | AmbP1, MAGNESIUM ION | | Authors: | Awakawa, T, Nakashima, Y, Mori, T, Abe, I. | | Deposit date: | 2018-01-10 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.361 Å) | | Cite: | Molecular Insight into the Mg2+-Dependent Allosteric Control of Indole Prenylation by Aromatic Prenyltransferase AmbP1
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5Z45
 
 | | Crystal structure of prenyltransferase AmbP1 pH6.5 complexed with GSPP and cis-indolyl vinyl isonitrile | | Descriptor: | 3-[(Z)-2-isocyanoethenyl]-1H-indole, AmbP1, GERANYL S-THIOLODIPHOSPHATE, ... | | Authors: | Awakawa, T, Nakashima, Y, Mori, T, Abe, I. | | Deposit date: | 2018-01-10 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Molecular Insight into the Mg2+-Dependent Allosteric Control of Indole Prenylation by Aromatic Prenyltransferase AmbP1
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5Z44
 
 | | Crystal structure of prenyltransferase AmbP1 complexed with GSPP | | Descriptor: | AmbP1, GERANYL S-THIOLODIPHOSPHATE, MAGNESIUM ION | | Authors: | Awakawa, T, Nakashima, Y, Mori, T, Abe, I. | | Deposit date: | 2018-01-10 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.458 Å) | | Cite: | Molecular Insight into the Mg2+-Dependent Allosteric Control of Indole Prenylation by Aromatic Prenyltransferase AmbP1
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5Z46
 
 | | Crystal structure of prenyltransferase AmbP1 pH8 complexed with GSPP and cis-indolyl vinyl isonitrile | | Descriptor: | 3-[(Z)-2-isocyanoethenyl]-1H-indole, AmbP1, GERANYL S-THIOLODIPHOSPHATE, ... | | Authors: | Awakawa, T, Nakashima, Y, Mori, T, Abe, I. | | Deposit date: | 2018-01-10 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Molecular Insight into the Mg2+-Dependent Allosteric Control of Indole Prenylation by Aromatic Prenyltransferase AmbP1
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
7VBR
 
 | |
7VBQ
 
 | | Heterodimer structure of Fe(II)/(alpha)ketoglutarate-dependent dioxygenase TlxIJ | | Descriptor: | FE (III) ION, Fe(II)/(alpha)ketoglutarate-dependent dioxygenase TlxI, Fe(II)/(alpha)ketoglutarate-dependent dioxygenase TlxJ, ... | | Authors: | Li, X, Awakawa, T, Mori, T, Abe, I. | | Deposit date: | 2021-09-01 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Heterodimeric Non-heme Iron Enzymes in Fungal Meroterpenoid Biosynthesis.
J.Am.Chem.Soc., 143, 2021
|
|
7EEH
 
 | | Selenomethionine labeled Fe(II)/(alpha)ketoglutarate-dependent dioxygenase TqaL | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FE (III) ION, TqaL | | Authors: | Bunno, R, Mori, T, Awakawa, T, Abe, I. | | Deposit date: | 2021-03-18 | | Release date: | 2021-05-26 | | Last modified: | 2021-07-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Aziridine Formation by a Fe II / alpha-Ketoglutarate Dependent Oxygenase and 2-Aminoisobutyrate Biosynthesis in Fungi.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
5Y4G
 
 | | Apo Structure of AmbP3 | | Descriptor: | AmbP3 | | Authors: | Wong, C.P, Awakawa, T, Nakashima, Y. | | Deposit date: | 2017-08-03 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two Distinct Substrate Binding Modes for the Normal and Reverse Prenylation of Hapalindoles by the Prenyltransferase AmbP3
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5Y72
 
 | | DMSPP Bound AmbP3 | | Descriptor: | AmbP3, DIMETHYLALLYL S-THIOLODIPHOSPHATE | | Authors: | Wong, C.P, Awakawa, T, Nakashima, Y. | | Deposit date: | 2017-08-16 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Two Distinct Substrate Binding Modes for the Normal and Reverse Prenylation of Hapalindoles by the Prenyltransferase AmbP3
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5Y84
 
 | | Hapalindole U and DMSPP Bound AmbP3 | | Descriptor: | AmbP3, DIMETHYLALLYL S-THIOLODIPHOSPHATE, Hapalindole U | | Authors: | Wong, C.P, Awakawa, T, Nakashima, Y. | | Deposit date: | 2017-08-18 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two Distinct Substrate Binding Modes for the Normal and Reverse Prenylation of Hapalindoles by the Prenyltransferase AmbP3
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5Y7C
 
 | | Hapalindole A and DMSPP Bound AmbP3 | | Descriptor: | AmbP3, DIMETHYLALLYL S-THIOLODIPHOSPHATE, Hapalindole A | | Authors: | Wong, C.P, Awakawa, T, Nakashima, Y. | | Deposit date: | 2017-08-16 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Two Distinct Substrate Binding Modes for the Normal and Reverse Prenylation of Hapalindoles by the Prenyltransferase AmbP3
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
4Y0K
 
 | |
7EXZ
 
 | | DgpB-DgpC complex apo 2.5 angstrom | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AP_endonuc_2 domain-containing protein, DgpB, ... | | Authors: | Mori, T, Senda, M, Senda, T, Abe, I. | | Deposit date: | 2021-05-29 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | C-Glycoside metabolism in the gut and in nature: Identification, characterization, structural analyses and distribution of C-C bond-cleaving enzymes.
Nat Commun, 12, 2021
|
|
7EXB
 
 | | DfgA-DfgB complex apo 2.4 angstrom | | Descriptor: | DfgB, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Mori, T, Senda, M, Senda, T, Abe, I. | | Deposit date: | 2021-05-26 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | C-Glycoside metabolism in the gut and in nature: Identification, characterization, structural analyses and distribution of C-C bond-cleaving enzymes.
Nat Commun, 12, 2021
|
|
5GK2
 
 | |
5GK0
 
 | |
5GK1
 
 | | Crystal structure of the ketosynthase StlD complexed with substrate | | Descriptor: | 3-OXO-5-METHYLHEXANOIC ACID, Ketosynthase StlD | | Authors: | Mori, T, Saito, Y, Morita, H, Abe, I. | | Deposit date: | 2016-07-03 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Insight into the Enzymatic Formation of Bacterial Stilbene.
Cell Chem Biol, 23, 2016
|
|
7BVR
 
 | | DgpB-DgpC complex apo | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AP_endonuc_2 domain-containing protein, DgpB, ... | | Authors: | Mori, T, He, H, Abe, I. | | Deposit date: | 2020-04-11 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | C-Glycoside metabolism in the gut and in nature: Identification, characterization, structural analyses and distribution of C-C bond-cleaving enzymes.
Nat Commun, 12, 2021
|
|
7BVS
 
 | | DfgA-DfgB complex apo | | Descriptor: | DfgB, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Mori, T, He, H, Abe, I. | | Deposit date: | 2020-04-11 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | C-Glycoside metabolism in the gut and in nature: Identification, characterization, structural analyses and distribution of C-C bond-cleaving enzymes.
Nat Commun, 12, 2021
|
|
7DRD
 
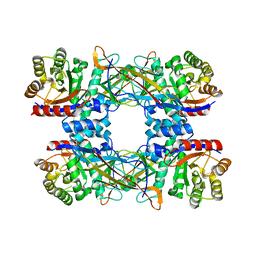 | | Cryo-EM structure of DgpB-C at 2.85 angstrom resolution | | Descriptor: | AP_endonuc_2 domain-containing protein, DgpB | | Authors: | Mori, T, Moriya, T, Adachi, N, Senda, T, Abe, I. | | Deposit date: | 2020-12-28 | | Release date: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | C-Glycoside metabolism in the gut and in nature: Identification, characterization, structural analyses and distribution of C-C bond-cleaving enzymes.
Nat Commun, 12, 2021
|
|
7DRE
 
 | | Cryo-EM structure of DfgA-B at 2.54 angstrom resolution | | Descriptor: | DfgB, Sugar phosphate isomerase/epimerase | | Authors: | Mori, T, Moriya, T, Adachi, N, Senda, T, Abe, I. | | Deposit date: | 2020-12-28 | | Release date: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | C-Glycoside metabolism in the gut and in nature: Identification, characterization, structural analyses and distribution of C-C bond-cleaving enzymes.
Nat Commun, 12, 2021
|
|
7DNM
 
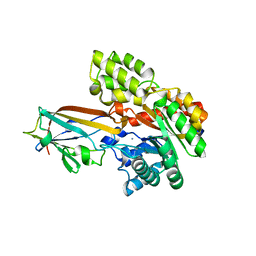 | | Crystal structure of the AgCarB2-C2 complex | | Descriptor: | AP_endonuc_2 domain-containing protein, AgCarC2, IODIDE ION, ... | | Authors: | Senda, M, Kumano, T, Watanabe, S, Kobayashi, M, Senda, T. | | Deposit date: | 2020-12-10 | | Release date: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the metabolism of xenobiotic C-glycosides by intestinal bacteria
Nat Commun, 2021
|
|
7DNN
 
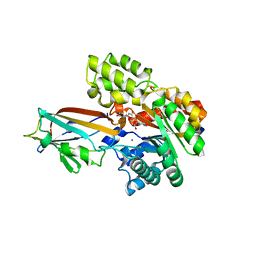 | | Crystal structure of the AgCarB2-C2 complex with homoorientin | | Descriptor: | 2-[3,4-bis(oxidanyl)phenyl]-6-[(2S,3R,4R,5S,6R)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]-5,7-bis(oxidanyl)chromen-4-one, AP_endonuc_2 domain-containing protein, AgCarC2, ... | | Authors: | Senda, M, Kumano, T, Watanabe, S, Kobayashi, M, Senda, T. | | Deposit date: | 2020-12-10 | | Release date: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for the metabolism of xenobiotic C-glycosides by intestinal bacteria
Nat Commun, 2021
|
|
4YZJ
 
 | |
