2WDS
 
 | | Crystal structure of the Streptomyces coelicolor H110A AcpS mutant in complex with cofactor CoA at 1.3 A | | Descriptor: | COENZYME A, HOLO-[ACYL-CARRIER-PROTEIN] SYNTHASE, MAGNESIUM ION | | Authors: | Dall'Aglio, P, Arthur, C, Crump, M.P, Crosby, J, Hadfield, A.T. | | Deposit date: | 2009-03-26 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Analysis of Streptomyces Coelicolor Phosphopantetheinyl Transferase, Acps, Reveals the Basis for Relaxed Substrate Specificity.
Biochemistry, 50, 2011
|
|
2WDY
 
 | | Crystal structure of the Streptomyces coelicolor D111A AcpS mutant in complex with cofactor CoA at 1.4 A | | Descriptor: | COENZYME A, HOLO-[ACYL-CARRIER-PROTEIN] SYNTHASE, MAGNESIUM ION, ... | | Authors: | Dall'Aglio, P, Arthur, C, Crump, M.P, Crosby, J, Hadfield, A.T. | | Deposit date: | 2009-03-27 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Analysis of Streptomyces Coelicolor Phosphopantetheinyl Transferase, Acps, Reveals the Basis for Relaxed Substrate Specificity.
Biochemistry, 50, 2011
|
|
2WDO
 
 | | Crystal structure of the S. coelicolor AcpS in complex with acetyl- CoA at 1.5 A | | Descriptor: | ACETYL COENZYME *A, COENZYME A, GLYCEROL, ... | | Authors: | Dall'Aglio, P, Arthur, C, Crump, M.P, Crosby, J, Hadfield, A.T. | | Deposit date: | 2009-03-25 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Analysis of Streptomyces Coelicolor Phosphopantetheinyl Transferase, Acps, Reveals the Basis for Relaxed Substrate Specificity.
Biochemistry, 50, 2011
|
|
3JC1
 
 | | Electron cryo-microscopy of the IST1-CHMP1B ESCRT-III copolymer | | Descriptor: | Charged multivesicular body protein 1b, Increased Sodium Tolerance 1 (IST1) | | Authors: | McCullough, J, Clippinger, A.K, Talledge, N, Skowyra, M.L, Saunders, M.G, Naismith, T.V, Colf, L.A, Afonine, P, Arthur, C, Sundquist, W.I, Hanson, P.I, Frost, A. | | Deposit date: | 2015-11-09 | | Release date: | 2015-12-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure and membrane remodeling activity of ESCRT-III helical polymers.
Science, 350, 2015
|
|
2JBZ
 
 | | Crystal structure of the Streptomyces coelicolor holo-[Acyl-carrier-protein] Synthase (AcpS) in complex with coenzyme A at 1.6 A | | Descriptor: | COENZYME A, HOLO-[ACYL-CARRIER-PROTEIN] SYNTHASE, MAGNESIUM ION, ... | | Authors: | Dall'Aglio, P, Arthur, C, Law, C.K.E, Crump, M.P, Crosby, J, Hadfield, A.T. | | Deposit date: | 2006-12-15 | | Release date: | 2007-01-23 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Analysis of Streptomyces Coelicolor Phosphopantetheinyl Transferase, Acps, Reveals the Basis for Relaxed Substrate Specificity.
Biochemistry, 50, 2011
|
|
2JCA
 
 | | Crystal structure of the streptomyces coelicolor holo- [Acyl-carrier-protein] Synthase (AcpS) at 2 A. | | Descriptor: | GLYCEROL, HOLO-[ACYL-CARRIER-PROTEIN] SYNTHASE, SODIUM ION, ... | | Authors: | Dall'Aglio, P, Arthur, C, Crump, M.P, Crosby, J, Hadfield, A.T. | | Deposit date: | 2006-12-21 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Analysis of Streptomyces Coelicolor Phosphopantetheinyl Transferase, Acps, Reveals the Basis for Relaxed Substrate Specificity.
Biochemistry, 50, 2011
|
|
7BDS
 
 | |
7BDR
 
 | | Structure of CTX-M-15 E166Q mutant crystallised in the presence of tazobactam (AAI101) | | Descriptor: | Beta-lactamase, CHLORIDE ION, SODIUM ION, ... | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2020-12-22 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.91 Å) | | Cite: | Penicillanic Acid Sulfones Inactivate the Extended-Spectrum beta-Lactamase CTX-M-15 through Formation of a Serine-Lysine Cross-Link: an Alternative Mechanism of beta-Lactamase Inhibition.
Mbio, 13, 2022
|
|
7SJO
 
 | | HtrA1S328A:Fab15H6.v4 complex | | Descriptor: | Fab15H6.v4 Heavy Chain, Fab15H6.v4 Light Chain, Serine protease HTRA1 | | Authors: | Gerhardy, S, Green, E, Estevez, A, Arthur, C.P, Ultsch, M, Rohou, A, Kirchhofer, D. | | Deposit date: | 2021-10-18 | | Release date: | 2022-09-07 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Allosteric inhibition of HTRA1 activity by a conformational lock mechanism to treat age-related macular degeneration.
Nat Commun, 13, 2022
|
|
7SJN
 
 | | HtrA1:Fab15H6.v4 complex | | Descriptor: | Fab15H6.v4 Heavy Chain, Fab15H6.v4 Light Chain, Serine protease HTRA1 | | Authors: | Gerhardy, S, Green, E, Estevez, A, Arthur, C.P, Ultsch, M, Rohou, A, Kirchhofer, D. | | Deposit date: | 2021-10-18 | | Release date: | 2022-09-07 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Allosteric inhibition of HTRA1 activity by a conformational lock mechanism to treat age-related macular degeneration.
Nat Commun, 13, 2022
|
|
7R3R
 
 | | Crystal structure of CTX-M-15 G238C mutant apoenzyme | | Descriptor: | Beta-lactamase, CHLORIDE ION, SULFATE ION | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2022-02-07 | | Release date: | 2022-05-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Penicillanic Acid Sulfones Inactivate the Extended-Spectrum beta-Lactamase CTX-M-15 through Formation of a Serine-Lysine Cross-Link: an Alternative Mechanism of beta-Lactamase Inhibition.
Mbio, 13, 2022
|
|
7QQC
 
 | | Structure of CTX-M-15 K73A mutant | | Descriptor: | Beta-lactamase, CHLORIDE ION, SULFATE ION | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2022-01-07 | | Release date: | 2022-05-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Penicillanic Acid Sulfones Inactivate the Extended-Spectrum beta-Lactamase CTX-M-15 through Formation of a Serine-Lysine Cross-Link: an Alternative Mechanism of beta-Lactamase Inhibition.
Mbio, 13, 2022
|
|
7QQ5
 
 | |
7R3Q
 
 | |
6Z7J
 
 | |
6Z7H
 
 | |
6Z7I
 
 | | Crystal structure of CTX-M-15 E166Q mutant apoenzyme | | Descriptor: | Beta-lactamase, GLYCEROL, SULFATE ION | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2020-05-31 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Penicillanic Acid Sulfones Inactivate the Extended-Spectrum beta-Lactamase CTX-M-15 through Formation of a Serine-Lysine Cross-Link: an Alternative Mechanism of beta-Lactamase Inhibition.
Mbio, 2022
|
|
6Z7K
 
 | | Crystal structure of CTX-M-15 in complex with the imine form of hydrolysed tazobactam | | Descriptor: | (2~{S},3~{S})-3-[bis(oxidanylidene)-$l^{5}-sulfanyl]-3-methyl-2-[(~{E})-3-oxidanylidenepropylideneamino]-4-(1,2,3-triaz ol-1-yl)butanoic acid, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2020-05-31 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Penicillanic Acid Sulfones Inactivate the Extended-Spectrum beta-Lactamase CTX-M-15 through Formation of a Serine-Lysine Cross-Link: an Alternative Mechanism of beta-Lactamase Inhibition.
Mbio, 2022
|
|
2KOS
 
 | |
7SJM
 
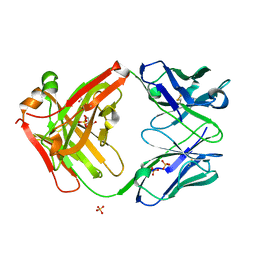 | | anti-HtrA1 Fab15H6.v4 | | Descriptor: | GLYCEROL, Heavy Chain, Light Chain, ... | | Authors: | Ultsch, M.H, Gerhardy, S. | | Deposit date: | 2021-10-18 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Allosteric inhibition of HTRA1 activity by a conformational lock mechanism to treat age-related macular degeneration.
Nat Commun, 13, 2022
|
|
7SJP
 
 | | anti-HtrA1 Fab15H6.v4 bound to HtrA1-LoopA peptide | | Descriptor: | GLYCEROL, Heavy Chain, HtrA1-LoopA peptide, ... | | Authors: | Ultsch, M.H, Gerhardy, S. | | Deposit date: | 2021-10-18 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Allosteric inhibition of HTRA1 activity by a conformational lock mechanism to treat age-related macular degeneration.
Nat Commun, 13, 2022
|
|
