6JK5
 
 | | Ca2+-dependent type II antifreeze protein (Ca2+-free form) | | Descriptor: | SULFATE ION, Type II antifreeze protein | | Authors: | Arai, T, Tsuda, S, Kondo, H, Nishimiya, Y. | | Deposit date: | 2019-02-27 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Calcium-Binding Generates the Semi-Clathrate Waters on a Type II Antifreeze Protein to Adsorb onto an Ice Crystal Surface.
Biomolecules, 9, 2019
|
|
6JK4
 
 | | Ca2+-dependent type II antifreeze protein | | Descriptor: | CALCIUM ION, Type II antifreeze protein | | Authors: | Arai, T, Tsuda, S, Kondo, H, Nishimiya, Y. | | Deposit date: | 2019-02-27 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Calcium-Binding Generates the Semi-Clathrate Waters on a Type II Antifreeze Protein to Adsorb onto an Ice Crystal Surface.
Biomolecules, 9, 2019
|
|
3ACS
 
 | |
4WD3
 
 | | Crystal structure of an L-amino acid ligase RizA | | Descriptor: | L-amino acid ligase | | Authors: | Kagawa, W, Arai, T, Kino, K, Kurumizaka, H. | | Deposit date: | 2014-09-06 | | Release date: | 2015-09-09 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of RizA, an L-amino-acid ligase from Bacillus subtilis.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
3OQJ
 
 | | Crystal structure of B. licheniformis CDPS yvmC-BLIC in complex with CAPSO | | Descriptor: | (2S)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, Putative uncharacterized protein yvmC | | Authors: | Bonnefond, L, Arai, T, Suzuki, T, Ishitani, R, Nureki, O. | | Deposit date: | 2010-09-03 | | Release date: | 2011-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structural basis for nonribosomal peptide synthesis by an aminoacyl-tRNA synthetase paralog.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3OQI
 
 | | Crystal structure of B. licheniformis CDPS yvmC-BLIC in complex with CHES | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Putative uncharacterized protein yvmC | | Authors: | Bonnefond, L, Arai, T, Suzuki, T, Ishitani, R, Nureki, O. | | Deposit date: | 2010-09-03 | | Release date: | 2011-02-23 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Structural basis for nonribosomal peptide synthesis by an aminoacyl-tRNA synthetase paralog.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3OQH
 
 | | Crystal structure of B. licheniformis CDPS yvmC-BLIC | | Descriptor: | GLYCEROL, Putative uncharacterized protein yvmC | | Authors: | Bonnefond, L, Arai, T, Suzuki, T, Ishitani, R, Nureki, O. | | Deposit date: | 2010-09-03 | | Release date: | 2011-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structural basis for nonribosomal peptide synthesis by an aminoacyl-tRNA synthetase paralog.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2Z1E
 
 | | Crystal structure of HypE from Thermococcus kodakaraensis (outward form) | | Descriptor: | Hydrogenase expression/formation protein HypE | | Authors: | Watanabe, S, Matsumi, R, Arai, T, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2007-05-08 | | Release date: | 2007-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structures of [NiFe] Hydrogenase Maturation Proteins HypC, HypD, and HypE: Insights into Cyanation Reaction by Thiol Redox Signaling
Mol.Cell, 27, 2007
|
|
2Z1C
 
 | | Crystal structure of HypC from Thermococcus kodakaraensis KOD1 | | Descriptor: | GLYCEROL, Hydrogenase expression/formation protein HypC, TETRAETHYLENE GLYCOL | | Authors: | Watanabe, S, Matsumi, R, Arai, T, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2007-05-08 | | Release date: | 2007-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of [NiFe] Hydrogenase Maturation Proteins HypC, HypD, and HypE: Insights into Cyanation Reaction by Thiol Redox Signaling
Mol.Cell, 27, 2007
|
|
2Z1F
 
 | | Crystal structure of HypE from Thermococcus kodakaraensis (inward form) | | Descriptor: | Hydrogenase expression/formation protein HypE | | Authors: | Watanabe, S, Matsumi, R, Arai, T, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2007-05-08 | | Release date: | 2007-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of [NiFe] Hydrogenase Maturation Proteins HypC, HypD, and HypE: Insights into Cyanation Reaction by Thiol Redox Signaling
Mol.Cell, 27, 2007
|
|
2Z1D
 
 | | Crystal structure of [NiFe] hydrogenase maturation protein, HypD from Thermococcus kodakaraensis | | Descriptor: | Hydrogenase expression/formation protein hypD, IRON/SULFUR CLUSTER | | Authors: | Watanabe, S, Matsumi, R, Arai, T, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2007-05-08 | | Release date: | 2007-07-17 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal Structures of [NiFe] Hydrogenase Maturation Proteins HypC, HypD, and HypE: Insights into Cyanation Reaction by Thiol Redox Signaling
Mol.Cell, 27, 2007
|
|
3A43
 
 | | Crystal structure of HypA | | Descriptor: | Hydrogenase nickel incorporation protein hypA, ZINC ION | | Authors: | Watanabe, S, Arai, T, Matsumi, R, Aromi, H, Imanaka, T, Miki, K. | | Deposit date: | 2009-06-30 | | Release date: | 2009-10-06 | | Last modified: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of HypA, a nickel-binding metallochaperone for [NiFe] hydrogenase maturation.
J.Mol.Biol., 394, 2009
|
|
7BWY
 
 | | Crystal structure of ice-binding protein from an Antarctic ascomycete, Antarctomyces psychrotrophicus. | | Descriptor: | GLYCEROL, Ice-binding protein isoform1a | | Authors: | Yamauchi, A, Arai, T, Kondo, H, Tsuda, S. | | Deposit date: | 2020-04-16 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | An Ice-Binding Protein from an Antarctic Ascomycete Is Fine-Tuned to Bind to Specific Water Molecules Located in the Ice Prism Planes.
Biomolecules, 10, 2020
|
|
7BWX
 
 | | Crystal structure of ice-binding protein from an Antarctic ascomycete, Antarctomyces psychrotrophicus. | | Descriptor: | GLYCEROL, Ice-binding protein isoform1a, SULFATE ION | | Authors: | Yamauchi, A, Arai, T, Kondo, H, Tsuda, S. | | Deposit date: | 2020-04-16 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | An Ice-Binding Protein from an Antarctic Ascomycete Is Fine-Tuned to Bind to Specific Water Molecules Located in the Ice Prism Planes.
Biomolecules, 10, 2020
|
|
7DC5
 
 | | Crystal structure of fungal antifreeze protein with intermediate activity | | Descriptor: | Antifreeze protein, SULFATE ION | | Authors: | Khan, N.M.M.U, Arai, T, Tsuda, S, Kondo, H. | | Deposit date: | 2020-10-23 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Characterization of microbial antifreeze protein with intermediate activity suggests that a bound-water network is essential for hyperactivity.
Sci Rep, 11, 2021
|
|
7DDB
 
 | | Crystal structure of fungal antifreeze protein with intermediate activity | | Descriptor: | Antifreeze protein, MAGNESIUM ION | | Authors: | Khan, N.M.M.U, Arai, T, Tsuda, S, Kondo, H. | | Deposit date: | 2020-10-28 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Characterization of microbial antifreeze protein with intermediate activity suggests that a bound-water network is essential for hyperactivity.
Sci Rep, 11, 2021
|
|
3A44
 
 | | Crystal structure of HypA in the dimeric form | | Descriptor: | Hydrogenase nickel incorporation protein hypA, ZINC ION | | Authors: | Watanabe, S, Arai, T, Matsumi, R, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2009-06-30 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Crystal structure of HypA, a nickel-binding metallochaperone for [NiFe] hydrogenase maturation.
J.Mol.Biol., 394, 2009
|
|
3VOT
 
 | | Crystal structure of L-amino acid ligase from Bacillus licheniformis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Suzuki, M, Takahashi, Y, Noguchi, A, Arai, T, Yagasaki, M, Kino, K, Saito, J. | | Deposit date: | 2012-02-08 | | Release date: | 2012-11-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of L-amino-acid ligase from Bacillus licheniformis
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3ACT
 
 | |
3AFJ
 
 | |
3S7T
 
 | |
5XQN
 
 | | Crystal structure of Notched-fin eelpout type III antifreeze protein (NFE6, AFP), C2221 form. | | Descriptor: | Ice-structuring protein, SULFATE ION | | Authors: | Adachi, M, Kondo, H, Tsuda, S. | | Deposit date: | 2017-06-07 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Polypentagonal ice-like water networks emerge solely in an activity-improved variant of ice-binding protein
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5XQR
 
 | | Crystal structure of Notched-fin eelpout type III antifreeze protein A20V mutant (NFE6, AFP), C2221 form | | Descriptor: | ACETATE ION, Ice-structuring protein | | Authors: | Adachi, M, Shimizu, R, Shibazaki, C, Kondo, H, Tsuda, S. | | Deposit date: | 2017-06-07 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Polypentagonal ice-like water networks emerge solely in an activity-improved variant of ice-binding protein
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5XQP
 
 | | Crystal structure of Notched-fin eelpout type III antifreeze protein (NFE6, AFP), P212121 form | | Descriptor: | Ice-structuring protein, SULFATE ION | | Authors: | Adachi, M, Shimizu, R, Shibazaki, C, Kondo, H, Tsuda, S. | | Deposit date: | 2017-06-07 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Polypentagonal ice-like water networks emerge solely in an activity-improved variant of ice-binding protein
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5XQU
 
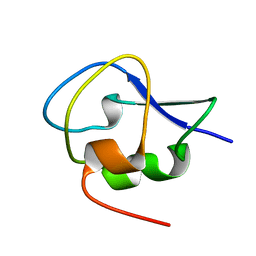 | | Crystal structure of Notched-fin eelpout type III antifreeze protein A20I mutant (NFE6, AFP), P212121 form | | Descriptor: | Ice-structuring protein | | Authors: | Adachi, M, Shimizu, R, Shibazaki, C, Kondo, H, Tsuda, S. | | Deposit date: | 2017-06-07 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Polypentagonal ice-like water networks emerge solely in an activity-improved variant of ice-binding protein
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
