1R2S
 
 | |
1R2T
 
 | |
1R2R
 
 | | CRYSTAL STRUCTURE OF RABBIT MUSCLE TRIOSEPHOSPHATE ISOMERASE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Aparicio, R, Ferreira, S.T, Polikarpov, I. | | Deposit date: | 2003-09-29 | | Release date: | 2003-12-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Closed conformation of the active site loop of rabbit muscle triosephosphate isomerase in the absence of substrate: evidence of conformational heterogeneity.
J.Mol.Biol., 334, 2003
|
|
3HZF
 
 | | Structure of TR-alfa bound to selective thyromimetic GC-1 in C2 space group | | Descriptor: | Thyroid hormone receptor, alpha isoform 1 variant, {4-[4-hydroxy-3-(1-methylethyl)benzyl]-3,5-dimethylphenoxy}acetic acid | | Authors: | Aparicio, R, Bleicher, L, Polikarpov, I. | | Deposit date: | 2009-06-23 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of GC-1 selectivity for thyroid hormone receptor isoforms.
Bmc Struct.Biol., 8, 2008
|
|
3ILZ
 
 | | Structure of TR-alfa bound to selective thyromimetic GC-1 in P212121 space group | | Descriptor: | Thyroid hormone receptor, alpha isoform 1 variant, {4-[4-hydroxy-3-(1-methylethyl)benzyl]-3,5-dimethylphenoxy}acetic acid | | Authors: | Aparicio, R, Polikarpov, L, Bleicher, L. | | Deposit date: | 2009-08-07 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of GC-1 selectivity for thyroid hormone receptor isoforms.
Bmc Struct.Biol., 8, 2008
|
|
5VOK
 
 | | Crystal structure of the C7orf59-HBXIP dimer | | Descriptor: | Ragulator complex protein LAMTOR4, Ragulator complex protein LAMTOR5 | | Authors: | Rasheed, N, Nascimento, A.F.Z, Bar-Peled, L, Shen, K, Sabatini, D.M, Aparicio, R, Smetana, J.H.C. | | Deposit date: | 2017-05-03 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | C7orf59/Lamtor4 phosphorylation and structural flexibility modulate Ragulator assembly.
Febs Open Bio, 2019
|
|
1YRR
 
 | | Crystal Structure Of The N-Acetylglucosamine-6-Phosphate Deacetylase From Escherichia Coli K12 at 2.0 A Resolution | | Descriptor: | GLYCEROL, N-acetylglucosamine-6-phosphate deacetylase, PHOSPHATE ION | | Authors: | Ferreira, F.M, Aparicio, R, Mendoza-Hernandez, G, Calcagno, M.L, Oliva, G. | | Deposit date: | 2005-02-04 | | Release date: | 2006-03-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of N-acetylglucosamine-6-phosphate deacetylase apoenzyme from Escherichia coli.
J.Mol.Biol., 359, 2006
|
|
4LNW
 
 | | Crystal structure of TR-alpha bound to T3 in a second site | | Descriptor: | 3,5,3'TRIIODOTHYRONINE, Thyroid hormone receptor alpha | | Authors: | Puhl, A.C, Aparicio, R, Polikarpov, I. | | Deposit date: | 2013-07-12 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of a new hormone-binding site on the surface of thyroid hormone receptor.
Mol.Endocrinol., 28, 2014
|
|
4LNX
 
 | | Crystal structure of TR-alpha bound to T4 in a second site | | Descriptor: | 3,5,3',5'-TETRAIODO-L-THYRONINE, 3,5,3'TRIIODOTHYRONINE, Thyroid hormone receptor alpha | | Authors: | Puhl, A.C, Aparicio, R, Polikarpov, I. | | Deposit date: | 2013-07-12 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Identification of a new hormone-binding site on the surface of thyroid hormone receptor.
Mol.Endocrinol., 28, 2014
|
|
4UOJ
 
 | | Structure of Fungal beta-mannosidase (GH2) from Trichoderma harzianum | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(3-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Muniz, J.R.C, Aparicio, R, Santos, J.C, Nascimento, A.S, Golubev, A.M, Polikarpov, I. | | Deposit date: | 2014-06-04 | | Release date: | 2014-07-09 | | Last modified: | 2023-03-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights Into the Structure and Function of Fungal Beta-Mannosidases from Glycoside Hydrolase Family 2 Based on Multiple Crystal Structures of the Trichoderma Harzianum Enzyme.
FEBS J., 281, 2014
|
|
2H77
 
 | | Crystal structure of human TR alpha bound T3 in monoclinic space group | | Descriptor: | 3,5,3'TRIIODOTHYRONINE, THRA protein | | Authors: | Nascimento, A.S, Dias, S.M.G, Nunes, F.M, Aparicio, R, Bleicher, L, Ambrosio, A.L.B, Figueira, A.C.M, Santos, M.A.M, Neto, M.O, Fischer, H, Togashi, H.F.M, Craievich, A.F, Garrat, R.C, Baxter, J.D, Webb, P, Polikarpov, I. | | Deposit date: | 2006-06-01 | | Release date: | 2006-07-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural rearrangements in the thyroid hormone receptor hinge domain and their putative role in the receptor function.
J.Mol.Biol., 360, 2006
|
|
2H79
 
 | | Crystal Structure of human TR alpha bound T3 in orthorhombic space group | | Descriptor: | 3,5,3'TRIIODOTHYRONINE, THRA protein | | Authors: | Nascimento, A.S, Dias, S.M.G, Nunes, F.M, Aparicio, R, Bleicher, L, Ambrosio, A.L.B, Figueira, A.C.M, Santos, M.A.M, Neto, M.O, Fischer, H, Togashi, H.F.M, Craievich, A.F, Garrat, R.C, Baxter, J.D, Webb, P, Polikarpov, I. | | Deposit date: | 2006-06-01 | | Release date: | 2006-07-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural rearrangements in the thyroid hormone receptor hinge domain and their putative role in the receptor function.
J.Mol.Biol., 360, 2006
|
|
5KSR
 
 | | Stationary phase survival protein E (SurE) from Xylella fastidiosa - XFSurE-TB (Tetramer Bigger). | | Descriptor: | 5'-nucleotidase SurE, CHLORIDE ION, IODIDE ION, ... | | Authors: | Machado, A.T.P, Fonseca, E.M.B, Dos Reis, M.A, Saraiva, A.M, Dos Santos, C.A, De Toledo, M.A, Polikarpov, I, De Souza, A.P, De Aparicio, R, Iulek, J. | | Deposit date: | 2016-07-09 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Conformational variability of the stationary phase survival protein E from Xylella fastidiosa revealed by X-ray crystallography, small-angle X-ray scattering studies, and normal mode analysis.
Proteins, 85, 2017
|
|
3IMY
 
 | |
5J7N
 
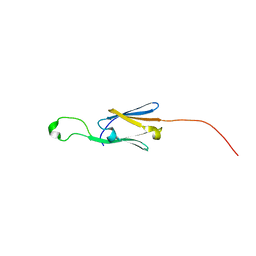 | | Crystal structure of a small heat-shock protein from Xylella fastidiosa reveals a distinct high order structure | | Descriptor: | Low molecular weight heat shock protein | | Authors: | Fonseca, E.M.B, Scorsato, V, dos Santos, C.A, Tomazini Jr, A, Aparicio, R, Polikarpov, I. | | Deposit date: | 2016-04-06 | | Release date: | 2017-04-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a small heat-shock protein from Xylella fastidiosa reveals a distinct high-order structure.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5JH2
 
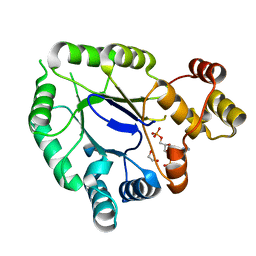 | | Crystal structure of the holo form of AKR4C7 from maize | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-2'-5'-DIPHOSPHATE, Aldose reductase, ... | | Authors: | Giuseppe, P.O, Santos, M.L, Sousa, S.M, Koch, K.E, Yunes, J.A, Aparicio, R, Murakami, M.T. | | Deposit date: | 2016-04-20 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | A comparative structural analysis reveals distinctive features of co-factor binding and substrate specificity in plant aldo-keto reductases.
Biochem.Biophys.Res.Commun., 474, 2016
|
|
5JGY
 
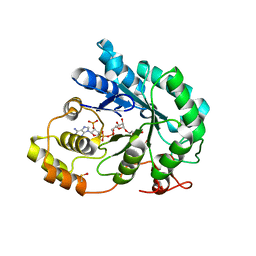 | | Crystal structure of maize AKR4C13 in P21 space group | | Descriptor: | 1,2-ETHANEDIOL, 4-{[(2R,3R,4S,5R)-5-({[(R)-{[(R)-{[(2R,3R,4R,5R)-5-(6-amino-9H-purin-9-yl)-3-hydroxy-4-(phosphonooxy)tetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}methyl)-3,4-dihydroxytetrahydrofuran-2-yl]oxy}butanoic acid (non-preferred name), Aldose reductase, ... | | Authors: | Santos, M.L, Giuseppe, P.O, Kiyota, E, Sousa, S.M, Schmelz, E.A, Yunes, J.A, Koch, K.E, Murakami, M.T, Aparicio, R. | | Deposit date: | 2016-04-20 | | Release date: | 2017-05-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of maize AKR4C13 in P21 space group
To Be Published
|
|
5JH1
 
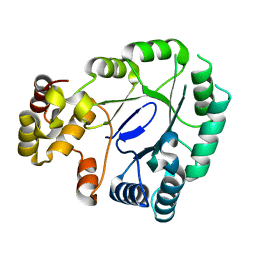 | | Crystal structure of the apo form of AKR4C7 from maize | | Descriptor: | Aldose reductase, AKR4C7 | | Authors: | Giuseppe, P.O, Santos, M.L, Sousa, S.M, Koch, K.E, Yunes, J.A, Aparicio, R, Murakami, M.T. | | Deposit date: | 2016-04-20 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A comparative structural analysis reveals distinctive features of co-factor binding and substrate specificity in plant aldo-keto reductases.
Biochem.Biophys.Res.Commun., 474, 2016
|
|
5JGW
 
 | | Crystal structure of maize AKR4C13 in complex with NADP and acetate | | Descriptor: | ACETATE ION, Aldose reductase, AKR4C13, ... | | Authors: | Santos, M.L, Giuseppe, P.O, Kiyota, E, Sousa, S.M, Schmelz, E.A, Yunes, J.A, Koch, K.E, Murakami, M.T, Aparicio, R. | | Deposit date: | 2016-04-20 | | Release date: | 2017-05-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of maize AKR4C13 in complex with NADP and acetate
To Be Published
|
|
5KSQ
 
 | | Stationary phase survival protein E (SurE) from Xylella fastidiosa | | Descriptor: | 5'-nucleotidase SurE, IODIDE ION, MANGANESE (II) ION, ... | | Authors: | Machado, A.T.P, Fonseca, E.M.B, Dos Reis, M.A, Saraiva, A.M, Dos Santos, C.A, De Toledo, M.A, Polikarpov, I, De Souza, A.P, Aparicio, R, Iulek, J. | | Deposit date: | 2016-07-09 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Conformational variability of the stationary phase survival protein E from Xylella fastidiosa revealed by X-ray crystallography, small-angle X-ray scattering studies, and normal mode analysis.
Proteins, 85, 2017
|
|
5KSS
 
 | | Stationary phase survival protein E (SurE) from Xylella fastidiosa - XFSurE-Ds (Dimer Smaller) | | Descriptor: | 5'-nucleotidase SurE, CHLORIDE ION, IODIDE ION, ... | | Authors: | Machado, A.T.P, Fonseca, E.M.B, Dos Reis, M.A, Saraiva, A.M, Dos Santos, C.A, De Toledo, A.M.S, Polikarpov, I, De Souza, A.P, Aparicio, R, Iulek, J. | | Deposit date: | 2016-07-09 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Conformational variability of the stationary phase survival protein E from Xylella fastidiosa revealed by X-ray crystallography, small-angle X-ray scattering studies, and normal mode analysis.
Proteins, 85, 2017
|
|
5KST
 
 | | Stationary phase Survival protein E (SurE) from Xylella fastidiosa- XfSurE-TSAmp (Tetramer Smaller - crystallization with 3'AMP). | | Descriptor: | 5'-nucleotidase SurE, IODIDE ION, MANGANESE (II) ION, ... | | Authors: | Machado, A.T.P, Fonseca, E.M.B, Dos Reis, M.A, Saraiva, A.M, Dos Santos, C.A, De Toledo, M.A.S, Polikarpov, I, De Souza, A.P, Aparicio, R, Iulek, J. | | Deposit date: | 2016-07-09 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.759 Å) | | Cite: | Conformational variability of the stationary phase survival protein E from Xylella fastidiosa revealed by X-ray crystallography, small-angle X-ray scattering studies, and normal mode analysis.
Proteins, 85, 2017
|
|
3GWS
 
 | | Crystal Structure of T3-Bound Thyroid Hormone Receptor | | Descriptor: | 3,5,3'TRIIODOTHYRONINE, Thyroid hormone receptor beta | | Authors: | Nascimento, A.S, Dias, S.M.G, Nunes, F.M, Aparicio, R, Polikarpov, I, Baxter, J.D, Webb, P. | | Deposit date: | 2009-04-01 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural rearrangements in the thyroid hormone receptor hinge domain and their putative role in the receptor function
J.Mol.Biol., 360, 2006
|
|
3JZB
 
 | | Crystal Structure of TR-alfa bound to the selective thyromimetic TRIAC | | Descriptor: | THRA protein, [4-(4-HYDROXY-3-IODO-PHENOXY)-3,5-DIIODO-PHENYL]-ACETIC ACID | | Authors: | Nascimento, A.S, Dias, S.M.G, Nunes, F.M, Aparicio, R. | | Deposit date: | 2009-09-23 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | Gaining ligand selectivity in thyroid hormone receptors via entropy.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3JZC
 
 | | Crystal Structure of TR-beta bound to the selective thyromimetic TRIAC | | Descriptor: | Thyroid hormone receptor beta, [4-(4-HYDROXY-3-IODO-PHENOXY)-3,5-DIIODO-PHENYL]-ACETIC ACID | | Authors: | Nascimento, A.S, Dias, S.G.M, Nunes, F.M, Aparicio, R. | | Deposit date: | 2009-09-23 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Gaining ligand selectivity in thyroid hormone receptors via entropy.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
