2WZ0
 
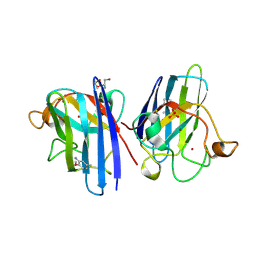 | | L38V SOD1 mutant complexed with aniline. | | Descriptor: | 2-METHOXY-5-METHYLANILINE, COPPER (II) ION, D-SERINE, ... | | Authors: | Antonyuk, S, Strange, R.W, Hasnain, S.S. | | Deposit date: | 2009-11-20 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural Discovery of Small Molecule Binding Sites in Cu-Zn Human Superoxide Dismutase Familial Amyotrophic Lateral Sclerosis Mutants Provides Insights for Lead Optimization.
J.Med.Chem., 53, 2010
|
|
6XY3
 
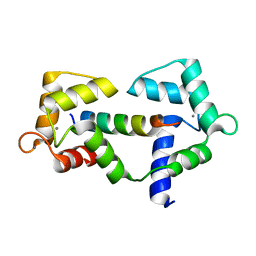 | |
6XXF
 
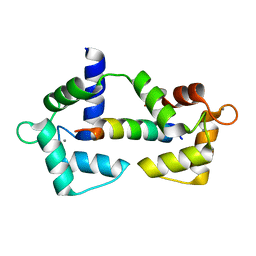 | |
6XXX
 
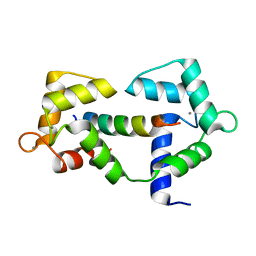 | | 1.25 Angstrom crystal structure of Ca/CaM A102V:RyR2 peptide complex | | Descriptor: | CALCIUM ION, Calmodulin-1, LYS-LYS-ALA-VAL-TRP-HIS-LYS-LEU-LEU-SER-LYS-GLN-ARG-LYS-ARG-ALA-VAL-VAL-ALA-CYS-PHE | | Authors: | Antonyuk, S, Helassa, N. | | Deposit date: | 2020-01-28 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | CPVT-associated calmodulin variants N53I and A102V dysregulate Ca2+ signalling via different mechanisms.
J.Cell.Sci., 135, 2022
|
|
2VR6
 
 | | Crystal Structure of G85R ALS mutant of Human Cu,Zn Superoxide Dismutase (CuZnSOD) at 1.3 A resolution | | Descriptor: | COPPER (II) ION, SULFATE ION, SUPEROXIDE DISMUTASE [CU-ZN], ... | | Authors: | Antonyuk, S, Cao, X, Seetharaman, S.V, Whitson, L.J, Taylor, A.B, Holloway, S.P, Strange, R.W, Doucette, P.A, Tiwari, A, Hayward, L.J, Padua, S, Cohlberg, J.A, Selverstone Valentine, J, Hasnain, S.S, Hart, P.J. | | Deposit date: | 2008-03-28 | | Release date: | 2008-04-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of the G85R Variant of Sod1 in Familial Amyotrophic Lateral Sclerosis.
J.Biol.Chem., 283, 2008
|
|
2VR8
 
 | | Crystal Structure of G85R ALS mutant of Human Cu,Zn Superoxide Dismutase (CuZnSOD) at 1.36 A resolution | | Descriptor: | COPPER (II) ION, SULFATE ION, SUPEROXIDE DISMUTASE [CU-ZN], ... | | Authors: | Antonyuk, S, Cao, X, Seetharaman, S.V, Whitson, L.J, Taylor, A.B, Holloway, S.P, Strange, R.W, Doucette, P.A, Tiwari, A, Hayward, L.J, Padua, S, Cohlberg, J.A, Selverstone Valentine, J, Hasnain, S.S, Hart, P.J. | | Deposit date: | 2008-03-28 | | Release date: | 2008-04-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structures of the G85R Variant of Sod1 in Familial Amyotrophic Lateral Sclerosis.
J.Biol.Chem., 283, 2008
|
|
2VR7
 
 | | Crystal Structure of G85R ALS mutant of Human Cu,Zn Superoxide Dismutase (CuZnSOD) at 1.58 A resolution | | Descriptor: | COPPER (II) ION, SULFATE ION, SUPEROXIDE DISMUTASE [CU-ZN], ... | | Authors: | Antonyuk, S, Cao, X, Seetharaman, S.V, Whitson, L.J, Taylor, A.B, Holloway, S.P, Strange, R.W, Doucette, P.A, Tiwari, A, Hayward, L.J, Padua, S, Cohlberg, J.A, Selverstone Valentine, J, Hasnain, S.S, Hart, P.J. | | Deposit date: | 2008-03-28 | | Release date: | 2008-04-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structures of the G85R Variant of Sod1 in Familial Amyotrophic Lateral Sclerosis.
J.Biol.Chem., 283, 2008
|
|
1GY2
 
 | | Crystal structure of Met148Leu rusticyanin | | Descriptor: | COPPER (II) ION, GLYCEROL, RUSTICYANIN | | Authors: | Hough, M.A, Kanbi, L.D, Antonyuk, S, Dodd, F, Hasnain, S. | | Deposit date: | 2002-04-16 | | Release date: | 2002-05-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal Structures of the met148Leu and Ser86Asp Mutants of Rusticyanin from Thiobacillus Ferrooxidans: Insights Into the Structural Relationship with the Cupredoxins and the Multi Copper Proteins.
J.Mol.Biol., 320, 2002
|
|
1GY1
 
 | | Crystal structures of Ser86Asp and Met148Leu Rusticyanin | | Descriptor: | COPPER (II) ION, RUSTICYANIN | | Authors: | Hough, M.A, Kanbi, L.D, Antonyuk, S, Dodd, F, Hasnain, S. | | Deposit date: | 2002-04-16 | | Release date: | 2002-05-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structures of the met148Leu and Ser86Asp Mutants of Rusticyanin from Thiobacillus Ferrooxidans: Insights Into the Structural Relationship with the Cupredoxins and the Multi Copper Proteins
J.Mol.Biol., 320, 2002
|
|
7R2U
 
 | |
7QQ2
 
 | |
7ZRQ
 
 | | 1.68 Angstrom crystal structure of Ca/CaM-E140G:CaMKIIdelta peptide complex | | Descriptor: | CALCIUM ION, Calcium/calmodulin-dependent protein kinase type II subunit delta, Calmodulin-1, ... | | Authors: | Helassa, N, Antonyuk, S. | | Deposit date: | 2022-05-04 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Calmodulin variant E140G associated with long QT syndrome impairs CaMKII delta autophosphorylation and L-type calcium channel inactivation.
J.Biol.Chem., 299, 2022
|
|
7ZRP
 
 | | 2.65 Angstrom crystal structure of Ca/CaM:CaMKIIdelta peptide complex | | Descriptor: | CALCIUM ION, Calcium/calmodulin-dependent protein kinase type II subunit delta, Calmodulin-1, ... | | Authors: | Helassa, N, Antonyuk, S. | | Deposit date: | 2022-05-04 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Calmodulin variant E140G associated with long QT syndrome impairs CaMKII delta autophosphorylation and L-type calcium channel inactivation.
J.Biol.Chem., 299, 2022
|
|
1HL5
 
 | | The Structure of Holo Type Human Cu, Zn Superoxide Dismutase | | Descriptor: | CALCIUM ION, COPPER (II) ION, SUPEROXIDE DISMUTASE, ... | | Authors: | Strange, R.W, Antonyuk, S, Hough, M.A, Doucette, P, Rodriguez, J, Hart, P.J, Hayward, L.J, Valentine, J.S, Hasnain, S.S. | | Deposit date: | 2003-03-13 | | Release date: | 2003-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of Holo and Metal-Deficient Wild-Type Human Cu, Zn Superoxide Dismutase and its Relevance to Familial Amyotrophic Lateral Sclerosis
J.Mol.Biol., 328, 2003
|
|
1HL4
 
 | | The Structure of Apo Type Human Cu, Zn Superoxide Dismutase | | Descriptor: | SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Strange, R.W, Antonyuk, S, Hough, M.A, Doucette, P, Rodriguez, J, Hart, P.J, Hayward, L.J, Valentine, J.S, Hasnain, S.S. | | Deposit date: | 2003-03-13 | | Release date: | 2003-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The Structure of Holo and Metal-Deficient Wild-Type Human Cu, Zn Superoxide Dismutase and its Relevance to Familial Amyotrophic Lateral Sclerosis
J.Mol.Biol., 328, 2003
|
|
1OEZ
 
 | | Zn His46Arg mutant of Human Cu, Zn Superoxide Dismutase | | Descriptor: | SULFATE ION, SUPEROXIDE DISMUTASE [CU-ZN], ZINC ION | | Authors: | Strange, R.W, Antonyuk, S, Hough, M.A, Doucette, P, Rodriguez, J, Elam, J.S, Hart, P.J, Hayward, L.J, Valentine, J.S, Hasnain, S.S. | | Deposit date: | 2003-04-02 | | Release date: | 2003-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Amyloid-Like Filaments and Water-Filled Nanotubes Formed by Sod1 Mutant Proteins Linked to Familial Als
Nat.Struct.Biol., 10, 2003
|
|
1OZU
 
 | | Crystal Structure of Familial ALS Mutant S134N of human Cu,Zn Superoxide Dismutase (CuZnSOD) to 1.3A resolution | | Descriptor: | SULFATE ION, Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Elam, J.S, Taylor, A.B, Strange, R, Antonyuk, S, Doucette, P.A, Rodriguez, J.A, Hasnain, S.S, Hayward, L.J, Valentine, J.S, Yeates, T.O, Hart, P.J. | | Deposit date: | 2003-04-09 | | Release date: | 2003-05-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Amyloid-like Filaments and Water-filled Nanotubes Formed by SOD1 Mutant Proteins Linked to Familial ALS
Nat.Struct.Biol., 10, 2003
|
|
1OZT
 
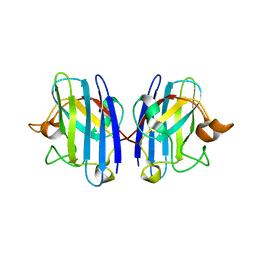 | | Crystal Structure of apo-H46R Familial ALS Mutant human Cu,Zn Superoxide Dismutase (CuZnSOD) to 2.5A resolution | | Descriptor: | Superoxide dismutase [Cu-Zn] | | Authors: | Elam, J.S, Taylor, A.B, Strange, R, Antonyuk, S, Doucette, P.A, Rodriguez, J.A, Hasnain, S.S, Hayward, L.J, Valentine, J.S, Yeates, T.O, Hart, P.J. | | Deposit date: | 2003-04-09 | | Release date: | 2003-05-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Amyloid-like Filaments and Water-filled Nanotubes Formed by SOD1 Mutant Proteins Linked to Familial ALS
Nat.Struct.Biol., 10, 2003
|
|
4CDA
 
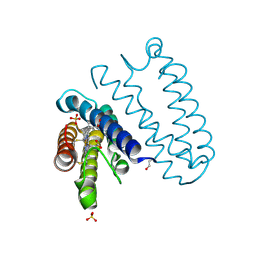 | | Spectroscopically-validated structure of ferric cytochrome c prime from Alcaligenes xylosoxidans | | Descriptor: | CYTOCHROME C', HEME C, SULFATE ION | | Authors: | Kekilli, D, Dworkowski, F, Antonyuk, S, Hough, M.A. | | Deposit date: | 2013-10-30 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Fingerprinting Redox and Ligand States in Haemprotein Crystal Structures Using Resonance Raman Spectroscopy.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4CDV
 
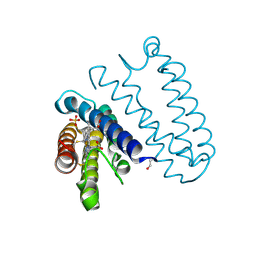 | | Spectroscopically-validated structure of cytochrome c prime from Alcaligenes xylosoxidans, reduced by X-ray irradiation at 100K | | Descriptor: | CYTOCHROME C', HEME C, SULFATE ION | | Authors: | Kekilli, D, Dworkowski, F, Antonyuk, S, Hough, M.A. | | Deposit date: | 2013-11-06 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Fingerprinting Redox and Ligand States in Haemprotein Crystal Structures Using Resonance Raman Spectroscopy.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4CDY
 
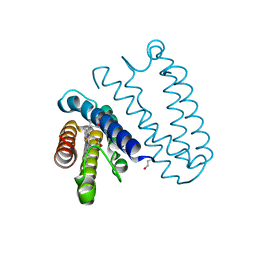 | | Spectroscopically-validated structure of cytochrome c prime from Alcaligenes xylosoxidans, reduced by X-ray irradiation at 160K | | Descriptor: | CYTOCHROME C', HEME C | | Authors: | Kekilli, D, Dworkowski, F, Antonyuk, S, Hough, M.A. | | Deposit date: | 2013-11-07 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Fingerprinting Redox and Ligand States in Haemprotein Crystal Structures Using Resonance Raman Spectroscopy.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4CJO
 
 | | Spectroscopically-validated structure of ferrous cytochrome c prime from Alcaligenes xylosoxidans, reduced at 180K using X-rays | | Descriptor: | CYTOCHROME C', HEME C | | Authors: | Kekilli, D, Dworkowski, F, Antonyuk, S, Hough, M.A. | | Deposit date: | 2013-12-21 | | Release date: | 2014-05-21 | | Last modified: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Fingerprinting Redox and Ligand States in Haemprotein Crystal Structures Using Resonance Raman Spectroscopy.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4CIP
 
 | | Spectroscopically-validated structure of ferrous cytochrome c prime from Alcaligenes xylosoxidans, reduced using ascorbate | | Descriptor: | ASCORBIC ACID, CYTOCHROME C', HEME C, ... | | Authors: | Kekilli, D, Dworkowski, F, Antonyuk, S, Hough, M.A. | | Deposit date: | 2013-12-13 | | Release date: | 2014-05-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Fingerprinting Redox and Ligand States in Haemprotein Crystal Structures Using Resonance Raman Spectroscopy.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4CJG
 
 | | Spectroscopically validated structure of the 5 coordinate proximal NO adduct of cytochrome c prime from Alcaligenes xylosoxidans | | Descriptor: | CYTOCHROME C', HEME C, NITRIC OXIDE | | Authors: | Kekilli, D, Dworkowski, F, Fuchs, M, Antonyuk, S, Hough, M.A. | | Deposit date: | 2013-12-20 | | Release date: | 2014-05-21 | | Last modified: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Fingerprinting Redox and Ligand States in Haemprotein Crystal Structures Using Resonance Raman Spectroscopy.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6FWF
 
 | | Low resolution structure of Neisseria meningitidis qNOR | | Descriptor: | CALCIUM ION, FE (III) ION, Nitric-oxide reductase, ... | | Authors: | Young, D, Antonyuk, S, Tosha, T, Hisano, T, Hasnain, S, Shiro, Y. | | Deposit date: | 2018-03-06 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Characterization of the quinol-dependent nitric oxide reductase from the pathogen Neisseria meningitidis, an electrogenic enzyme.
Sci Rep, 8, 2018
|
|
