1WPA
 
 | |
1XAW
 
 | |
1KWA
 
 | | HUMAN CASK/LIN-2 PDZ DOMAIN | | Descriptor: | HCASK/LIN-2 PROTEIN, SULFATE ION | | Authors: | Daniels, D.L, Cohen, A.R, Anderson, J.M, Brunger, A.T. | | Deposit date: | 1998-01-16 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of the hCASK PDZ domain reveals the structural basis of class II PDZ domain target recognition
Nat.Struct.Biol., 5, 1998
|
|
4YYX
 
 | |
2QUO
 
 | |
2RCZ
 
 | | Structure of the second PDZ domain of ZO-1 | | Descriptor: | Tight junction protein ZO-1 | | Authors: | Lavie, A, Lye, M.F. | | Deposit date: | 2007-09-20 | | Release date: | 2007-10-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Domain swapping within PDZ2 is responsible for dimerization of ZO proteins.
J.Biol.Chem., 282, 2007
|
|
2MDU
 
 | | Circular Permutant of the WW Domain with Loop 1 Excised | | Descriptor: | Pin1 WW domain | | Authors: | Kier, B.L. | | Deposit date: | 2013-09-18 | | Release date: | 2014-01-15 | | Last modified: | 2014-01-29 | | Method: | SOLUTION NMR | | Cite: | Circular Permutation of a WW Domain: Folding Still Occurs after Excising the Turn of the Folding-Nucleating Hairpin.
J.Am.Chem.Soc., 136, 2014
|
|
3TSZ
 
 | |
3TSW
 
 | | crystal structure of the PDZ3-SH3-GUK core module of Human ZO-1 | | Descriptor: | SULFATE ION, Tight junction protein ZO-1 | | Authors: | Nomme, J, Lavie, A. | | Deposit date: | 2011-09-13 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.847 Å) | | Cite: | The Src Homology 3 Domain Is Required for Junctional Adhesion Molecule Binding to the Third PDZ Domain of the Scaffolding Protein ZO-1.
J.Biol.Chem., 286, 2011
|
|
3TSV
 
 | |
4OEP
 
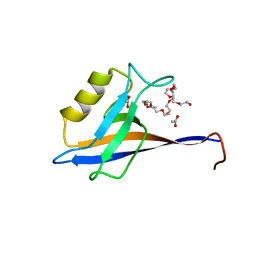 | |
4OEO
 
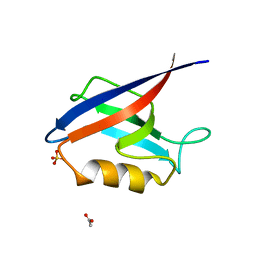 | |
1BFE
 
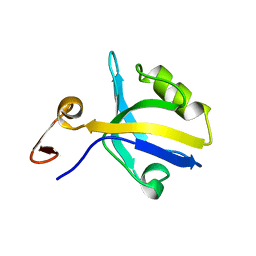 | | THE THIRD PDZ DOMAIN FROM THE SYNAPTIC PROTEIN PSD-95 | | Descriptor: | PSD-95 | | Authors: | Doyle, D.A, Lee, A, Lewis, J, Kim, E, Sheng, M, Mackinnon, R. | | Deposit date: | 1998-05-20 | | Release date: | 1998-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ.
Cell(Cambridge,Mass.), 85, 1996
|
|
1BE9
 
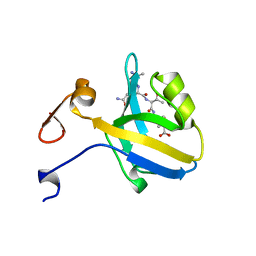 | | THE THIRD PDZ DOMAIN FROM THE SYNAPTIC PROTEIN PSD-95 IN COMPLEX WITH A C-TERMINAL PEPTIDE DERIVED FROM CRIPT. | | Descriptor: | CRIPT, PSD-95 | | Authors: | Doyle, D.A, Lee, A, Lewis, J, Kim, E, Sheng, M, Mackinnon, R. | | Deposit date: | 1998-05-20 | | Release date: | 1998-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ.
Cell(Cambridge,Mass.), 85, 1996
|
|
3LH5
 
 | |
1G9O
 
 | | FIRST PDZ DOMAIN OF THE HUMAN NA+/H+ EXCHANGER REGULATORY FACTOR | | Descriptor: | NHE-RF | | Authors: | Karthikeyan, S, Leung, T, Birrane, G, Webster, G, Ladias, J.A.A. | | Deposit date: | 2000-11-26 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the PDZ1 domain of human Na(+)/H(+) exchanger regulatory factor provides insights into the mechanism of carboxyl-terminal leucine recognition by class I PDZ domains.
J.Mol.Biol., 308, 2001
|
|
