1LV0
 
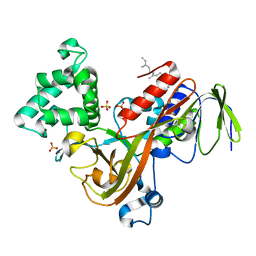 | | Crystal structure of the Rab effector guanine nucleotide dissociation inhibitor (GDI) in complex with a geranylgeranyl (GG) peptide | | Descriptor: | GERAN-8-YL GERAN, RAB GDP disossociation inhibitor alpha, SULFATE ION | | Authors: | An, Y, Shao, Y, Alory, C, Matteson, J, Sakisaka, T, Chen, W, Gibbs, R.A, Wilson, I.A, Balch, W.E. | | Deposit date: | 2002-05-23 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Geranylgeranyl switching regulates GDI-Rab GTPase recycling.
Structure, 11, 2003
|
|
8IM8
 
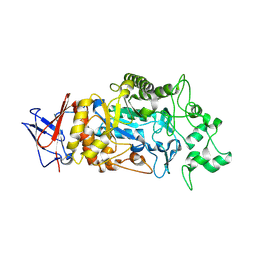 | | Crystal structure of Periplasmic alpha-amylase (MalS) from E.coli | | Descriptor: | CALCIUM ION, Periplasmic alpha-amylase | | Authors: | An, Y, Park, J.T, Park, K.H, Woo, E.J. | | Deposit date: | 2023-03-06 | | Release date: | 2023-05-24 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Distinctive Permutated Domain Structure of Periplasmic alpha-Amylase (MalS) from Glycoside Hydrolase Family 13 Subfamily 19.
Molecules, 28, 2023
|
|
3GRL
 
 | |
3GQ2
 
 | |
7VFT
 
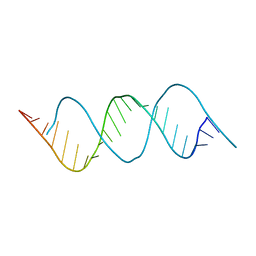 | | Crystal structure of rGGGC(CAG)5GUCC oligo | | Descriptor: | RNA (5'-R(*GP*GP*GP*CP*CP*AP*GP*CP*AP*GP*CP*AP*GP*CP*AP*GP*CP*AP*GP*GP*UP*CP*C)-3') | | Authors: | An, Y, Chan, H.Y.E, Ngo, J.C.K. | | Deposit date: | 2021-09-13 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular insights into the interaction of CAG trinucleotide RNA repeats with nucleolin and its implication in polyglutamine diseases.
Nucleic Acids Res., 50, 2022
|
|
4UW2
 
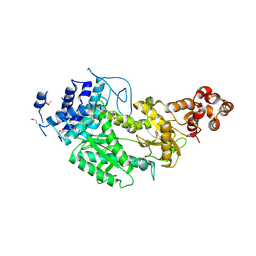 | | Crystal structure of Csm1 in T.onnurineus | | Descriptor: | CSM1 | | Authors: | Jung, T.Y, An, Y, Park, K.H, Lee, M.H, Oh, B.H, Woo, E.J. | | Deposit date: | 2014-08-08 | | Release date: | 2015-03-25 | | Last modified: | 2015-09-23 | | Method: | X-RAY DIFFRACTION (2.632 Å) | | Cite: | Crystal Structure of the Csm1 Subunit of the Csm Complex and its Single-Stranded DNA-Specific Nuclease Activity.
Structure, 23, 2015
|
|
2NW4
 
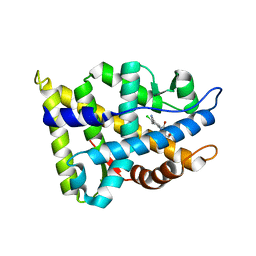 | | Crystal Structure of the Rat Androgen Receptor Ligand Binding Domain Complex with BMS-564929 | | Descriptor: | 2-CHLORO-4-[(7R,7AS)-7-HYDROXY-1,3-DIOXOTETRAHYDRO-1H-PYRROLO[1,2-C]IMIDAZOL-2(3H)-YL]-3-METHYLBENZONITRILE, Androgen receptor | | Authors: | Ostrowski, J, Kuhns, J.E, Lupisella, J.A, Manfredi, M.C, Beehler, B.C, Krystek, S.R, Bi, Y, Sun, C, Seethala, R, Golla, R, Sleph, P.G, Fura, A, An, Y, Kish, K.F, Sack, J.S, Mookhtiar, K.A, Grover, G.J, Hamann, L.G. | | Deposit date: | 2006-11-14 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Pharmacological and x-ray structural characterization of a novel selective androgen receptor modulator: potent hyperanabolic stimulation of skeletal muscle with hypostimulation of prostate in rats.
Endocrinology, 148, 2007
|
|
4RQU
 
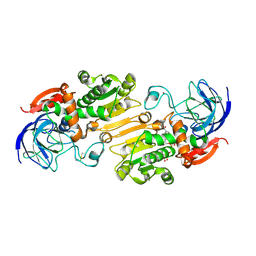 | | Alcohol Dehydrogenase crystal structure in complex with NAD | | Descriptor: | Alcohol Dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Xu, Y.W. | | Deposit date: | 2014-11-05 | | Release date: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insight into the conformational change of alcohol dehydrogenase from arabidopsis Thalianal during coenzyme binding.
Biochimie, 108C, 2014
|
|
4RQT
 
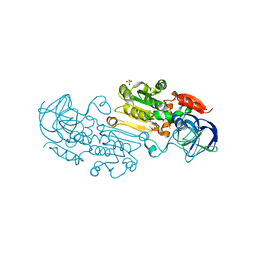 | | Alcohol Dehydrogenase Crystal Structure | | Descriptor: | ACETIC ACID, Alcohol dehydrogenase class-P, SULFATE ION, ... | | Authors: | Xu, Y.W. | | Deposit date: | 2014-11-05 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insight into the conformational change of alcohol dehydrogenase from arabidopsis Thalianal during coenzyme binding.
Biochimie, 108C, 2014
|
|
3KDU
 
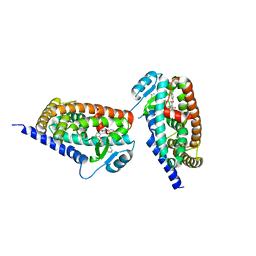 | |
3KDT
 
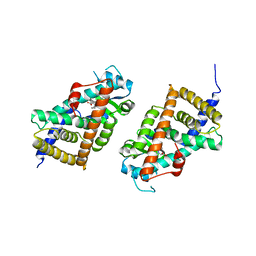 | |
4XHD
 
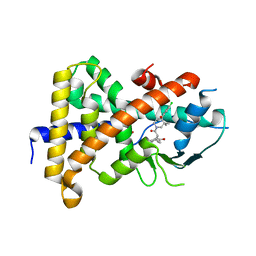 | | STRUCTURE OF HUMAN PREGNANE X RECEPTOR LIGAND BINDING DOMAIN WITH COMPOUND-1 | | Descriptor: | GLYCEROL, N-{(2R)-1-[(4S)-4-(4-chlorophenyl)-4-hydroxy-3,3-dimethylpiperidin-1-yl]-3-methyl-1-oxobutan-2-yl}-2-cyclopropylacetamide, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Khan, J.A, Camac, D.M. | | Deposit date: | 2015-01-05 | | Release date: | 2015-01-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Developing Adnectins That Target SRC Co-Activator Binding to PXR: A Structural Approach toward Understanding Promiscuity of PXR.
J.Mol.Biol., 427, 2015
|
|
4S0T
 
 | |
4S0S
 
 | |
3CE3
 
 | |
3G0W
 
 | |
2IHQ
 
 | |
7C02
 
 | | Crystal structure of dimeric MERS-CoV receptor binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Dai, L, Qi, J, Gao, G.F. | | Deposit date: | 2020-04-30 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | A Universal Design of Betacoronavirus Vaccines against COVID-19, MERS, and SARS.
Cell, 182, 2020
|
|
3F82
 
 | |
6P9F
 
 | | Crystal structure of RAR-related orphan receptor C (NHIS-RORGT(244-487)-L6-SRC1(678-692)) in complex with a phenyl (3-phenylpyrrolidin-3-yl)sulfone inhibitor | | Descriptor: | Nuclear receptor ROR-gamma, Nuclear receptor coactivator 1 chimera, trans-4-{(3R)-3-[(4-fluorophenyl)sulfonyl]-3-[4-(1,1,1,2,3,3,3-heptafluoropropan-2-yl)phenyl]pyrrolidine-1-carbonyl}cyclohexane-1-carboxylic acid | | Authors: | Sack, J. | | Deposit date: | 2019-06-10 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of potent, selective and orally bioavailable phenyl ((R)-3-phenylpyrrolidin-3-yl)sulfone analogues as ROR gamma t inverse agonists.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
3CTH
 
 | |
3CTJ
 
 | |
6ITU
 
 | | Crystal Structure of the GULP1 PTB domain-APP peptide complex | | Descriptor: | 1,2-ETHANEDIOL, Amyloid beta A4 protein, PTB domain-containing engulfment adapter protein 1, ... | | Authors: | Yung, K.W.Y, Lau, K.F, Ngo, J.C.K. | | Deposit date: | 2018-11-26 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Attenuation of amyloid-beta generation by atypical protein kinase C-mediated phosphorylation of engulfment adaptor PTB domain containing 1 threonine 35.
Faseb J., 33, 2019
|
|
5IHL
 
 | |
8DNT
 
 | | SARS-CoV-2 specific T cell receptor | | Descriptor: | Beta-2-microglobulin, MHC class I antigen alpha chain, Nucleoprotein, ... | | Authors: | Gallagher, D.T, Wu, D, Gowthaman, R, Pierce, B.G, Mariuzza, R.A, Weng, N.P. | | Deposit date: | 2022-07-11 | | Release date: | 2023-07-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | SARS-CoV-2 infection establishes a stable and age-independent CD8 + T cell response against a dominant nucleocapsid epitope using restricted T cell receptors.
Nat Commun, 14, 2023
|
|
