4V0B
 
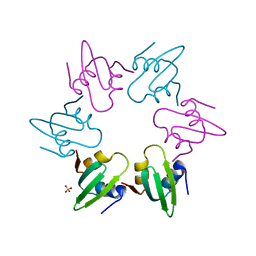 | | Escherichia coli FtsH hexameric N-domain | | Descriptor: | ATP-DEPENDENT ZINC METALLOPROTEASE FTSH, SULFATE ION | | Authors: | Serek-Heuberger, J, Martin, J, Lupas, A.N, Hartmann, M.D. | | Deposit date: | 2014-09-13 | | Release date: | 2015-01-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure and Evolution of N-Domains in Aaa Metalloproteases.
J.Mol.Biol., 427, 2015
|
|
5ONB
 
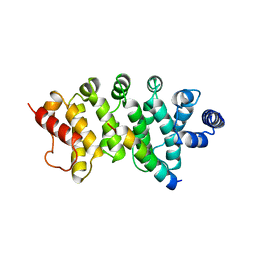 | | Drosophila Bag-of-marbles CBM peptide bound to human CAF40 | | Descriptor: | CCR4-NOT transcription complex subunit 9, GLYCEROL, Protein bag-of-marbles | | Authors: | Raisch, T, Sgromo, A, Backhaus, C, Izaurralde, E, Weichenrieder, O. | | Deposit date: | 2017-08-03 | | Release date: | 2017-12-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | DrosophilaBag-of-marbles directly interacts with the CAF40 subunit of the CCR4-NOT complex to elicit repression of mRNA targets.
RNA, 24, 2018
|
|
8AE1
 
 | |
6GDW
 
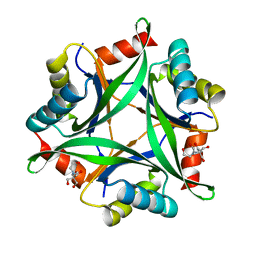 | |
6GDV
 
 | | Structure of CutA from Synechococcus elongatus PCC7942 complexed with Bis-Tris molecule | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Periplasmic divalent cation tolerance protein | | Authors: | Tremino, L, Rubio, V. | | Deposit date: | 2018-04-24 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and structural characterization of PII-like protein CutA does not support involvement in heavy metal tolerance and hints at a small-molecule carrying/signaling role.
Febs J., 2020
|
|
6GDX
 
 | |
6GDU
 
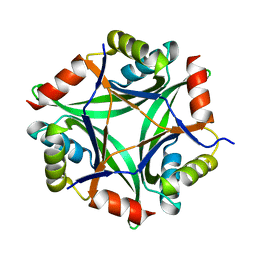 | | Structure of CutA from Synechococcus elongatus PCC7942 | | Descriptor: | Periplasmic divalent cation tolerance protein | | Authors: | Tremino, L, Rubio, V. | | Deposit date: | 2018-04-24 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Functional and structural characterization of PII-like protein CutA does not support involvement in heavy metal tolerance and hints at a small-molecule carrying/signaling role.
Febs J., 2020
|
|
2MUY
 
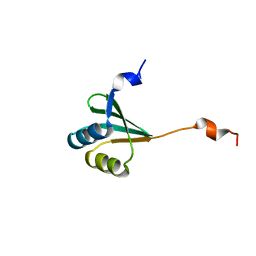 | | The solution structure of the FtsH periplasmic N-domain | | Descriptor: | ATP-dependent zinc metalloprotease FtsH | | Authors: | Scharfenberg, F, Serek-Heuberger, J, Martin, J, Lupas, A.N, Coles, M. | | Deposit date: | 2014-09-18 | | Release date: | 2015-01-28 | | Last modified: | 2015-02-25 | | Method: | SOLUTION NMR | | Cite: | Structure and Evolution of N-domains in AAA Metalloproteases.
J.Mol.Biol., 427, 2015
|
|
8CIO
 
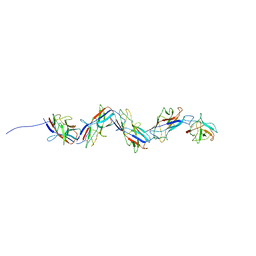 | |
8CKA
 
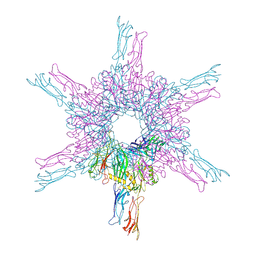 | | Deinococcus radidurans HPI S-layer | | Descriptor: | Hexagonally packed intermediate-layer surface protein | | Authors: | von Kuegelgen, A, Yamashita, K, Murshudov, G, Bharat, T. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-19 | | Last modified: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Interdigitated immunoglobulin arrays form the hyperstable surface layer of the extremophilic bacterium Deinococcus radiodurans.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
2P3M
 
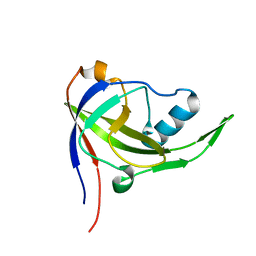 | | Solution structure of Mj0056 | | Descriptor: | Riboflavin Kinase MJ0056 | | Authors: | Coles, M, Truffault, V, Djuranovic, S, Martin, J, Lupas, A.N. | | Deposit date: | 2007-03-09 | | Release date: | 2007-10-09 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | A CTP-Dependent Archaeal Riboflavin Kinase Forms a Bridge in the Evolution of Cradle-Loop Barrels.
Structure, 15, 2007
|
|
2VBV
 
 | | Riboflavin kinase Mj0056 from Methanocaldococcus jannaschii in complex with CDP and FMN | | Descriptor: | CHLORIDE ION, CYTIDINE-5'-DIPHOSPHATE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Hartmann, M.D, Ammelburg, M, Djuranovic, S, Martin, J, Lupas, A.N, Zeth, K. | | Deposit date: | 2007-09-16 | | Release date: | 2007-11-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Ctp-Dependent Archaeal Riboflavin Kinase Forms a Bridge in the Evolution of Cradle-Loop Barrels.
Structure, 15, 2007
|
|
2VBU
 
 | | Riboflavin kinase Mj0056 from Methanocaldococcus jannaschii in complex with CDP | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CYTIDINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Hartmann, M.D, Ammelburg, M, Djuranovic, S, Martin, J, Lupas, A.N, Zeth, K. | | Deposit date: | 2007-09-16 | | Release date: | 2007-11-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Ctp-Dependent Archaeal Riboflavin Kinase Forms a Bridge in the Evolution of Cradle-Loop Barrels.
Structure, 15, 2007
|
|
2VBS
 
 | | Riboflavin kinase Mj0056 from Methanocaldococcus jannaschii in complex with PO4 | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, RIBOFLAVIN KINASE, ... | | Authors: | Hartmann, M.D, Djuranovic, S, Ammelburg, M, Martin, J, Lupas, A.N, Zeth, K. | | Deposit date: | 2007-09-16 | | Release date: | 2007-11-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Ctp-Dependent Archaeal Riboflavin Kinase Forms a Bridge in the Evolution of Cradle-Loop Barrels.
Structure, 15, 2007
|
|
2VBT
 
 | | Riboflavin kinase Mj0056 from Methanocaldococcus jannaschii in complex with CDP and PO4 | | Descriptor: | CYTIDINE-5'-DIPHOSPHATE, PHOSPHATE ION, RIBOFLAVIN KINASE, ... | | Authors: | Hartmann, M.D, Ammelburg, M, Djuranovic, S, Martin, J, Lupas, A.N, Zeth, K. | | Deposit date: | 2007-09-16 | | Release date: | 2007-11-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Ctp-Dependent Archaeal Riboflavin Kinase Forms a Bridge in the Evolution of Cradle-Loop Barrels.
Structure, 15, 2007
|
|
6T76
 
 | | PII-like protein CutA from Nostoc sp. PCC 7120 in apo form | | Descriptor: | Periplasmic divalent cation tolerance protein, SULFATE ION | | Authors: | Selim, K.A, Albrecht, R, Forchhammer, K, Hartmann, M.D. | | Deposit date: | 2019-10-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Functional and structural characterization of PII-like protein CutA does not support involvement in heavy metal tolerance and hints at a small-molecule carrying/signaling role.
Febs J., 288, 2021
|
|
6T7E
 
 | | PII-like protein CutA from Nostoc sp. PCC7120 in complex with MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Periplasmic divalent cation tolerance protein | | Authors: | Selim, K.A, Albrecht, R, Forchhammer, K, Hartmann, M.D. | | Deposit date: | 2019-10-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Functional and structural characterization of PII-like protein CutA does not support involvement in heavy metal tolerance and hints at a small-molecule carrying/signaling role.
Febs J., 288, 2021
|
|
4C46
 
 | | ANDREI-N-LVPAS fused to GCN4 adaptors | | Descriptor: | BROMIDE ION, GENERAL CONTROL PROTEIN GCN4 | | Authors: | Albrecht, R, Alva, V, Ammelburg, M, Baer, K, Basina, E, Boichenko, I, Bonhoeffer, F, Braun, V, Chaubey, M, Chauhan, N, Chellamuthu, V.R, Coles, M, Deiss, S, Ewers, C.P, Forouzan, D, Fuchs, A, Groemping, Y, Hartmann, M.D, Hernandez Alvarez, B, Jeganantham, A, Kalev, I, Koenninger, U, Koiwai, K, Kopec, K.O, Korycinski, M, Laudenbach, B, Lehmann, K, Leo, J.C, Linke, D, Marialke, J, Martin, J, Mechelke, M, Michalik, M, Noll, A, Patzer, S.I, Scharfenberg, F, Schueckel, M, Shahid, S.A, Sulz, E, Ursinus, A, Wuertenberger, S, Zhu, H. | | Deposit date: | 2013-08-30 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Your Personalized Protein Structure: Andrei N. Lupas Fused to GCN4 Adaptors.
J.Struct.Biol., 186, 2014
|
|
5LOX
 
 | |
5LOY
 
 | |
5ONA
 
 | | Drosophila Bag-of-marbles CBM peptide bound to human CAF40-CNOT1 | | Descriptor: | CCR4-NOT transcription complex subunit 1, CCR4-NOT transcription complex subunit 9, CHLORIDE ION, ... | | Authors: | Raisch, T, Sgromo, A, Backhaus, C, Izaurralde, E, Weichenrieder, O. | | Deposit date: | 2017-08-03 | | Release date: | 2017-12-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | DrosophilaBag-of-marbles directly interacts with the CAF40 subunit of the CCR4-NOT complex to elicit repression of mRNA targets.
RNA, 24, 2018
|
|
7PTR
 
 | |
7PTT
 
 | |
7PTP
 
 | |
7PTU
 
 | |
