7B7S
 
 | | CDK2/cyclin A2 in complex with 3H-pyrazolo[4,3-f]quinoline-based derivative HSD1368 | | Descriptor: | 7-(3-(trifluoromethyl)-1H-pyrazol-4yl)-3,8,10,11-tetrahydropyrazolo[4,3-f]thiopyrano[3,4-c]quinoline 9-oxide, Cyclin-A2, Cyclin-dependent kinase 2, ... | | Authors: | Djukic, S, Skerlova, J, Rezacova, P. | | Deposit date: | 2020-12-11 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | 3 H -Pyrazolo[4,3- f ]quinoline-Based Kinase Inhibitors Inhibit the Proliferation of Acute Myeloid Leukemia Cells In Vivo.
J.Med.Chem., 64, 2021
|
|
1ANG
 
 | | CRYSTAL STRUCTURE OF HUMAN ANGIOGENIN REVEALS THE STRUCTURAL BASIS FOR ITS FUNCTIONAL DIVERGENCE FROM RIBONUCLEASE | | Descriptor: | ANGIOGENIN | | Authors: | Acharya, K.R, Allen, S, Shapiro, R, Riordan, J.F, Vallee, B.L. | | Deposit date: | 1994-01-18 | | Release date: | 1995-04-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human angiogenin reveals the structural basis for its functional divergence from ribonuclease.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
1HE7
 
 | | Human Nerve growth factor receptor TrkA | | Descriptor: | GLYCEROL, HIGH AFFINITY NERVE GROWTH FACTOR RECEPTOR | | Authors: | Banfield, M, Robertson, A, Allen, S, Dando, J, Tyler, S, Bennett, G, Brain, S, Mason, G, Holden, P, Clarke, A, Naylor, R, Wilcock, G, Brady, R, Dawbarn, D. | | Deposit date: | 2000-11-20 | | Release date: | 2001-04-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification and Structure of the Nerve Growth Factor Binding Site on Trka.
Biochem.Biophys.Res.Commun., 282, 2001
|
|
3B50
 
 | | Structure of H. influenzae sialic acid binding protein bound to Neu5Ac. | | Descriptor: | N-acetyl-beta-neuraminic acid, Sialic acid-binding periplasmic protein siaP | | Authors: | Ramaswamy, S, Coussens, N.P, Apicella, M.A, Johnston, J.W. | | Deposit date: | 2007-10-25 | | Release date: | 2007-11-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Characterization of the N-Acetyl-5-neuraminic Acid-binding Site of the Extracytoplasmic Solute Receptor (SiaP) of Nontypeable Haemophilus influenzae Strain 2019
J.Biol.Chem., 283, 2008
|
|
5UBY
 
 | | Fab structure of anti-HIV-1 gp120 mAb 1A8 | | Descriptor: | Heavy chain of Fab fragment of anti-HIV1 gp120 mAb 1A8, Light chain of Fab fragment of anti-HIV1 gp120 mAb 1A8 | | Authors: | Pan, R, Kong, X.-P. | | Deposit date: | 2016-12-21 | | Release date: | 2018-01-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | High Antibody Diversity and Low Inter-clonal Competition Favor Production of Functional Neutralizing Antibodies
To be Published
|
|
5UBZ
 
 | | Fab structure of HIV gp120 specific mAb 1E12 | | Descriptor: | Anti-HIV1 gp120 mAb 1E12 Fab heavy chain, Anti-HIV1 gp120 mAb 1E12 Fab light chain | | Authors: | Pan, R, Kong, X.-P. | | Deposit date: | 2016-12-21 | | Release date: | 2018-01-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | High Antibody Diversity and Low Inter-clonal Competition Favor Production of Functional Neutralizing Antibodies
To be published
|
|
6X3L
 
 | | Sortilin-Progranulin Interaction With Compound 2 | | Descriptor: | 1-benzyl-3-tert-butyl-1H-pyrazole-5-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Parthasarathy, G, Soisson, S.M. | | Deposit date: | 2020-05-21 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of potent inhibitors of the sortilin-progranulin interaction.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6X48
 
 | | Sortilin-Progranulin Interaction With Compound 17 | | Descriptor: | GLYCEROL, N-(3,5-dichlorobenzene-1-carbonyl)-5,5-dimethyl-L-norleucine, Sortilin, ... | | Authors: | Parthasarathy, G, Soisson, S.M. | | Deposit date: | 2020-05-22 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification of potent inhibitors of the sortilin-progranulin interaction.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6X4H
 
 | | Sortilin-Progranulin Interaction With Compound 24 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-methyl-N-(6-phenoxypyridine-3-carbonyl)-L-leucine, GLYCEROL, ... | | Authors: | Parthasarathy, G, Soisson, S.M, Klein, D. | | Deposit date: | 2020-05-22 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification of potent inhibitors of the sortilin-progranulin interaction.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
7RT5
 
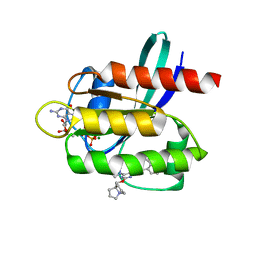 | | Crystal structure of KRAS G12D with compound 36 (4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-7-(8-ethynyl-7-fluoronaphthalen-1-yl)-8-fluoro-2-{[(4s,7as)-tetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}pyrido[4,3-d]pyrimidine) bound | | Descriptor: | 4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-7-(8-ethynyl-7-fluoronaphthalen-1-yl)-8-fluoro-2-{[(4s,7as)-tetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}pyrido[4,3-d]pyrimidine, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Gunn, R.J, Thomas, N.C, Xiaolun, W, Lawson, J.D, Marx, M.A. | | Deposit date: | 2021-08-12 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Identification of MRTX1133, a Noncovalent, Potent, and Selective KRAS G12D Inhibitor.
J.Med.Chem., 65, 2022
|
|
7RT4
 
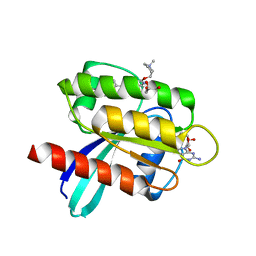 | | KRAS G12D in complex with Compound 5B (7-(8-chloronaphthalen-1-yl)-8-fluoro-2-{[(2S)-1-methylpyrrolidin-2-yl]methoxy}-4-(piperazin-1-yl)pyrido[4,3-d]pyrimidine) | | Descriptor: | 7-(8-chloronaphthalen-1-yl)-8-fluoro-2-{[(2S)-1-methylpyrrolidin-2-yl]methoxy}-4-(piperazin-1-yl)pyrido[4,3-d]pyrimidine, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gunn, R.J, Thomas, N.C, Xiaolun, W, Lawson, J.D, Marx, M.A. | | Deposit date: | 2021-08-12 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of MRTX1133, a Noncovalent, Potent, and Selective KRAS G12D Inhibitor.
J.Med.Chem., 65, 2022
|
|
7RPZ
 
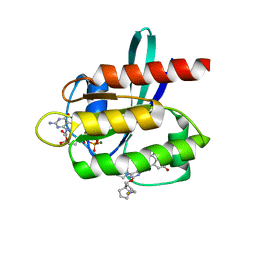 | | KRAS G12D in complex with MRTX-1133 | | Descriptor: | 4-(4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2R,4R,7aS)-2-fluorotetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}pyrido[4,3-d]pyrimidin-7-yl)-5-ethynyl-6-fluoronaphthalen-2-ol, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Gunn, R.J, Thomas, N.C, Xiaolun, W, Lawson, J.D, Marx, M.A. | | Deposit date: | 2021-08-05 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Identification of MRTX1133, a Noncovalent, Potent, and Selective KRAS G12D Inhibitor.
J.Med.Chem., 65, 2022
|
|
7RT1
 
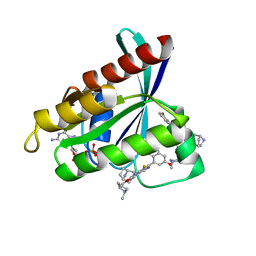 | | Crystal Structure of KRAS G12D with compound 15 (4-(4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2S)-1-methylpyrrolidin-2-yl]methoxy}pyrido[4,3-d]pyrimidin-7-yl)naphthalen-2-ol) bound | | Descriptor: | 4-(4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2S)-1-methylpyrrolidin-2-yl]methoxy}pyrido[4,3-d]pyrimidin-7-yl)naphthalen-2-ol, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Gunn, R.J, Thomas, N.C, Xiaolun, W, Lawson, J.D, Marx, M.A. | | Deposit date: | 2021-08-12 | | Release date: | 2021-12-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Identification of MRTX1133, a Noncovalent, Potent, and Selective KRAS G12D Inhibitor.
J.Med.Chem., 65, 2022
|
|
7RT2
 
 | | Crystal Structure of KRAS G12D with compound 25 (4-(4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2R,4R,7aS)-2-fluorotetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}pyrido[4,3-d]pyrimidin-7-yl)naphthalen-2-ol) bound | | Descriptor: | 1,2-ETHANEDIOL, 4-(4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2R,4R,7aS)-2-fluorotetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}pyrido[4,3-d]pyrimidin-7-yl)naphthalen-2-ol, GLYCEROL, ... | | Authors: | Gunn, R.J, Thomas, N.C, Xiaolun, W, Lawson, J.D, Marx, M.A. | | Deposit date: | 2021-08-12 | | Release date: | 2021-12-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Identification of MRTX1133, a Noncovalent, Potent, and Selective KRAS G12D Inhibitor.
J.Med.Chem., 65, 2022
|
|
7RT3
 
 | | Crystal Structure of KRAS G12D with compound 24 (4-(4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2S,4S,7aR)-2-fluorotetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}pyrido[4,3-d]pyrimidin-7-yl)naphthalen-2-ol) bound | | Descriptor: | 4-(4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2S,4S,7aR)-2-fluorotetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}pyrido[4,3-d]pyrimidin-7-yl)naphthalen-2-ol, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Gunn, R.J, Thomas, N.C, Xiaolun, W, Lawson, J.D, Marx, M.A. | | Deposit date: | 2021-08-12 | | Release date: | 2021-12-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Identification of MRTX1133, a Noncovalent, Potent, and Selective KRAS G12D Inhibitor.
J.Med.Chem., 65, 2022
|
|
4MMP
 
 | |
4MNP
 
 | |
