7D80
 
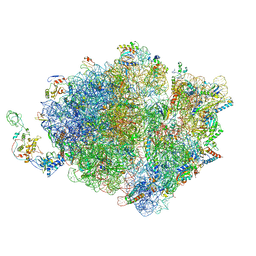 | | Molecular model of the cryo-EM structure of 70S ribosome in complex with peptide deformylase, trigger factor, and methionine aminopeptidase | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Akbar, S, Bhakta, S, Sengupta, J. | | Deposit date: | 2020-10-06 | | Release date: | 2021-04-07 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insights into the interplay of protein biogenesis factors with the 70S ribosome.
Structure, 29, 2021
|
|
7D6Z
 
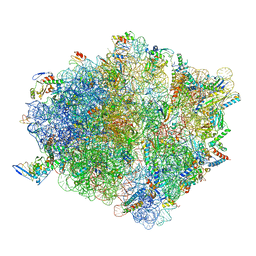 | | Molecular model of the cryo-EM structure of 70S ribosome in complex with peptide deformylase and trigger factor | | Descriptor: | 16S ribosomal rRNA, 23S ribosomal rRNA, 30S ribosomal protein S10, ... | | Authors: | Akbar, S, Bhakta, S, Sengupta, J. | | Deposit date: | 2020-10-02 | | Release date: | 2021-04-07 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into the interplay of protein biogenesis factors with the 70S ribosome.
Structure, 29, 2021
|
|
6IY7
 
 | |
6J45
 
 | |
6IZ7
 
 | |
6IZI
 
 | |
6J0A
 
 | |
