1DT1
 
 | | THERMUS THERMOPHILUS CYTOCHROME C552 SYNTHESIZED BY ESCHERICHIA COLI | | Descriptor: | CYTOCHROME C552, HEME C | | Authors: | Fee, J.A, Chen, Y, Hill, M.J, Gomez-Moran, E, Loehr, T, Ai, J, Thony-Meyer, L, Williams, P.A, Stura, E, Sridhar, V, McRee, D.E. | | Deposit date: | 2000-01-10 | | Release date: | 2000-02-18 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Integrity of thermus thermophilus cytochrome c552 synthesized by Escherichia coli cells expressing the host-specific cytochrome c maturation genes, ccmABCDEFGH: biochemical, spectral, and structural characterization of the recombinant protein.
Protein Sci., 9, 2000
|
|
1R0Q
 
 | | Characterization of the conversion of the malformed, recombinant cytochrome rc552 to a 2-formyl-4-vinyl (Spirographis) heme | | Descriptor: | 2-FORMYL-PROTOPORPHRYN IX, Cytochrome c-552 | | Authors: | Fee, J.A, Todaro, T.R, Luna, E, Sanders, D, Hunsicker-Wang, L.M, Patel, K.M, Bren, K.L, Gomez-Moran, E, Hill, M.G, Ai, J, Loehr, T.M, Oertling, W.A, Williams, P.A, Stout, C.D, McRee, D, Pastuszyn, A. | | Deposit date: | 2003-09-22 | | Release date: | 2004-09-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Cytochrome rC552, formed during expression of the truncated, Thermus thermophilus cytochrome c552 gene in the cytoplasm of Escherichia coli, reacts spontaneously to form protein-bound 2-formyl-4-vinyl (Spirographis) heme.
Biochemistry, 43, 2004
|
|
1QYZ
 
 | | Characterization of the malformed, recombinant cytochrome rC552 | | Descriptor: | 2-ACETYL-PROTOPORPHYRIN IX, Cytochrome c-552 | | Authors: | Fee, J.A, Todaro, T.R, Luna, E, Sanders, D, Hunsicker-Wang, L.M, Patel, K.M, Bren, K.L, Gomez-Moran, E, Hill, M.G, Ai, J, Loehr, T.M, Oertling, W.A, Williams, P.A, Stout, C.D, McRee, D, Pastuszyn, A. | | Deposit date: | 2003-09-12 | | Release date: | 2004-09-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Cytochrome rC552, formed during expression of the truncated, Thermus thermophilus cytochrome c552 gene in the cytoplasm of Escherichia coli, reacts spontaneously to form protein-bound 2-formyl-4-vinyl (Spirographis) heme.
Biochemistry, 43, 2004
|
|
1I4Y
 
 | | THE CRYSTAL STRUCTURE OF PHASCOLOPSIS GOULDII WILD TYPE METHEMERYTHRIN | | Descriptor: | CHLORIDE ION, METHEMERYTHRIN, MU-OXO-DIIRON | | Authors: | Farmer, C.S, Kurtz Jr, D.M, Liu, Z.-J, Wang, B.C, Rose, J. | | Deposit date: | 2001-02-23 | | Release date: | 2001-03-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structures of Phascolopsis gouldii wild type and L98Y methemerythrins: structural and functional alterations of the O2 binding pocket.
J.Biol.Inorg.Chem., 6, 2001
|
|
1I4Z
 
 | | THE CRYSTAL STRUCTURE OF PHASCOLOPSIS GOULDII L98Y METHEMERYTHRIN | | Descriptor: | METHEMERYTHRIN, MU-OXO-DIIRON | | Authors: | Farmer, C.S, Kurtz Jr, D.M, Liu, Z.-J, Wang, B.C, Rose, J. | | Deposit date: | 2001-02-23 | | Release date: | 2001-03-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structures of Phascolopsis gouldii wild type and L98Y methemerythrins: structural and functional alterations of the O2 binding pocket.
J.Biol.Inorg.Chem., 6, 2001
|
|
4ZSA
 
 | | Crystal structure of FGFR1 kinase domain in complex with 7n | | Descriptor: | 4-(4-ethylpiperazin-1-yl)-N-[6-(3-methoxyphenyl)-2H-indazol-3-yl]benzamide, Fibroblast growth factor receptor 1 | | Authors: | Liu, Q.F, Xu, Y.C. | | Deposit date: | 2015-05-13 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, synthesis and biological evaluation of novel FGFR inhibitors bearing an indazole scaffold.
Org.Biomol.Chem., 13, 2015
|
|
5Z0S
 
 | | Crystal structure of FGFR1 kinase domain in complex with a novel inhibitor | | Descriptor: | 1-[(6-chloroimidazo[1,2-b]pyridazin-3-yl)sulfonyl]-6-(1-methyl-1H-pyrazol-4-yl)-1H-pyrazolo[4,3-b]pyridine, Fibroblast growth factor receptor 1 | | Authors: | Liu, Q, Xu, Y. | | Deposit date: | 2017-12-20 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-Based Discovery of a Series of 5H-Pyrrolo[2,3-b]pyrazine FGFR Kinase Inhibitors
Molecules, 23, 2018
|
|
5EOB
 
 | | Crystal structure of CMET in complex with novel inhibitor | | Descriptor: | 6-[bis(fluoranyl)-[6-(4-fluorophenyl)-[1,2,4]triazolo[4,3-b][1,2,4]triazin-3-yl]methyl]quinoline, Hepatocyte growth factor receptor | | Authors: | Liu, Q, Chen, T, Xu, Y. | | Deposit date: | 2015-11-10 | | Release date: | 2016-10-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of 6-(difluoro(6-(4-fluorophenyl)-[1,2,4]triazolo[4,3-b][1,2,4]triazin-3-yl)methyl)quinoline as a highly potent and selective c-Met inhibitor
Eur.J.Med.Chem., 116, 2016
|
|
6IUO
 
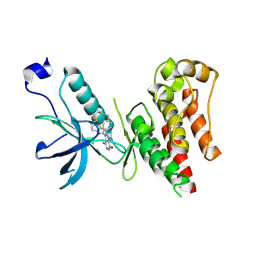 | | Crystal structure of FGFR4 kinase domain in complex with a covalent inhibitor | | Descriptor: | Fibroblast growth factor receptor 4, N-({4-[4-amino-3-(3,5-dimethyl-1-benzofuran-2-yl)-7-oxo-6,7-dihydro-2H-pyrazolo[3,4-d]pyridazin-2-yl]phenyl}methyl)prop-2-enamide | | Authors: | Xu, Y, Liu, Q. | | Deposit date: | 2018-11-29 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and Development of a Series of Pyrazolo[3,4-d]pyridazinone Compounds as the Novel Covalent Fibroblast Growth Factor Receptor Inhibitors by the Rational Drug Design.
J.Med.Chem., 62, 2019
|
|
4MXC
 
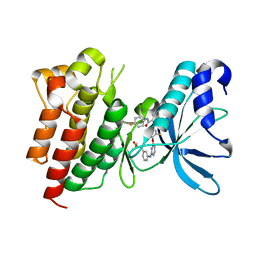 | | Crystal structure of CMET in complex with novel inhibitor | | Descriptor: | Hepatocyte growth factor receptor, N-(3-fluoro-4-{[2-({3-[(methylsulfonyl)methyl]phenyl}amino)pyrimidin-4-yl]oxy}phenyl)-N'-(4-fluorophenyl)cyclopropane-1,1-dicarboxamide | | Authors: | Liu, Q.F, Chen, T.T, Xu, Y.C. | | Deposit date: | 2013-09-26 | | Release date: | 2014-10-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.632 Å) | | Cite: | Discovery of Anilinopyrimidines as Dual Inhibitors of c-Met and VEGFR-2: Synthesis, SAR, and Cellular Activity
ACS MED.CHEM.LETT., 5, 2014
|
|
4GG5
 
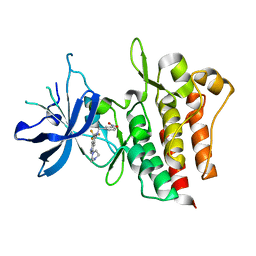 | | Crystal structure of CMET in complex with novel inhibitor | | Descriptor: | 3-(4-methylpiperazin-1-yl)-N-(3-nitrobenzyl)-7-(trifluoromethyl)quinolin-5-amine, Hepatocyte growth factor receptor | | Authors: | Liu, Q.F, Chen, T.T, Xu, Y.C. | | Deposit date: | 2012-08-05 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.423 Å) | | Cite: | Multisubstituted quinoxalines and pyrido[2,3-d]pyrimidines: Synthesis and SAR study as tyrosine kinase c-Met inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4GG7
 
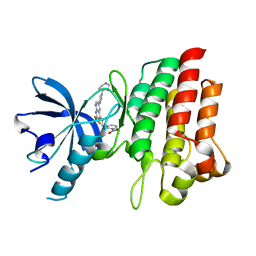 | | Crystal structure of cMET in complex with novel inhibitor | | Descriptor: | Hepatocyte growth factor receptor, N-(3-nitrobenzyl)-6-[1-(piperidin-4-yl)-1H-pyrazol-4-yl]-2-(trifluoromethyl)pyrido[2,3-d]pyrimidin-4-amine | | Authors: | Liu, Q.F, Chen, T.T, Xu, Y.C. | | Deposit date: | 2012-08-06 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Multisubstituted quinoxalines and pyrido[2,3-d]pyrimidines: Synthesis and SAR study as tyrosine kinase c-Met inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6ITJ
 
 | | Crystal structure of FGFR1 kinase domain in complex with compound 3 | | Descriptor: | 4-azanyl-3-(3,5-dimethyl-1-benzofuran-2-yl)-2-phenyl-6~{H}-pyrazolo[3,4-d]pyridazin-7-one, Fibroblast growth factor receptor 1 | | Authors: | Xu, Y, Liu, Q. | | Deposit date: | 2018-11-23 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | Discovery and Development of a Series of Pyrazolo[3,4-d]pyridazinone Compounds as the Novel Covalent Fibroblast Growth Factor Receptor Inhibitors by the Rational Drug Design.
J.Med.Chem., 62, 2019
|
|
6IUP
 
 | | Crystal structure of FGFR4 kinase domain in complex with compound 5 | | Descriptor: | DIMETHYL SULFOXIDE, Fibroblast growth factor receptor 4, N-{4-[4-amino-3-(3,5-dimethyl-1-benzofuran-2-yl)-7-oxo-6,7-dihydro-2H-pyrazolo[3,4-d]pyridazin-2-yl]phenyl}prop-2-enamide | | Authors: | Xu, Y, Liu, Q. | | Deposit date: | 2018-11-29 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and Development of a Series of Pyrazolo[3,4-d]pyridazinone Compounds as the Novel Covalent Fibroblast Growth Factor Receptor Inhibitors by the Rational Drug Design.
J.Med.Chem., 62, 2019
|
|
7V3S
 
 | | Crystal structure of CMET in complex with a novel inhibitor | | Descriptor: | Hepatocyte growth factor receptor, ~{N}1'-[3-fluoranyl-4-(10~{H}-pyrido[3,2-b][1,4]benzoxazin-4-yloxy)phenyl]-~{N}1-(4-fluorophenyl)cyclopropane-1,1-dicarboxamide | | Authors: | Su, H.X, Liu, Q.F, Chen, T.T, Li, M.J, Xu, Y.C. | | Deposit date: | 2021-08-11 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of 10H-Benzo[b]pyrido[2,3-e][1,4]oxazine AXL Inhibitors via Structure-Based Drug Design Targeting c-Met Kinase
J.Med.Chem., 66, 2023
|
|
7V3R
 
 | | Crystal structure of CMET in complex with a novel inhibitor | | Descriptor: | Hepatocyte growth factor receptor, ~{N}1'-[3-fluoranyl-4-(2-phenylazanylpyrimidin-4-yl)oxy-phenyl]-~{N}1-(4-fluorophenyl)cyclopropane-1,1-dicarboxamide | | Authors: | Su, H.X, Liu, Q.F, Chen, T.T, Li, M.J, Xu, Y.C. | | Deposit date: | 2021-08-11 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of 10H-Benzo[b]pyrido[2,3-e][1,4]oxazine AXL Inhibitors via Structure-Based Drug Design Targeting c-Met Kinase
J.Med.Chem., 66, 2023
|
|
