7ABB
 
 | |
7ABA
 
 | |
6ZSU
 
 | |
6I86
 
 | | Crocagin biosynthetic gene J | | Descriptor: | GLYCEROL, Uncharacterized protein | | Authors: | Adam, S, Koehnke, J. | | Deposit date: | 2018-11-19 | | Release date: | 2019-03-13 | | Last modified: | 2019-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of CgnJ, a domain of unknown function protein from the crocagin gene cluster.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
8A2N
 
 | | Structure of crocagin biosynthetic protein CgnD | | Descriptor: | CgnD, SULFATE ION | | Authors: | Adam, S, Koehnke, J. | | Deposit date: | 2022-06-06 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Unusual peptide-binding proteins guide pyrroloindoline alkaloid formation in crocagin biosynthesis.
Nat.Chem., 15, 2023
|
|
8A9C
 
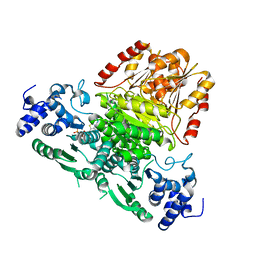 | |
8AGY
 
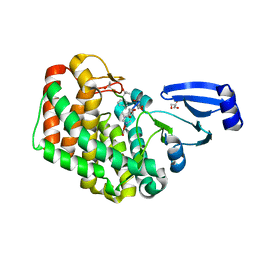 | |
8AHD
 
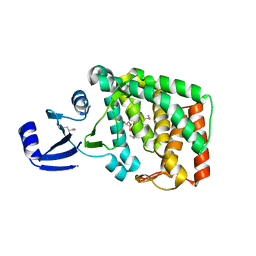 | |
8A8Y
 
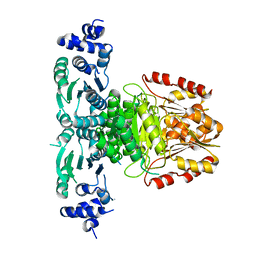 | |
6Z8L
 
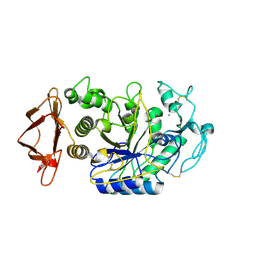 | | Alpha-Amylase in complex with probe fragments | | Descriptor: | CALCIUM ION, CHLORIDE ION, Pancreatic alpha-amylase, ... | | Authors: | Adam, S, Koehnke, J. | | Deposit date: | 2020-06-02 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.40000367 Å) | | Cite: | Enhancing glycan stability via site-selective fluorination: modulating substrate orientation by molecular design.
Chem Sci, 12, 2020
|
|
6TET
 
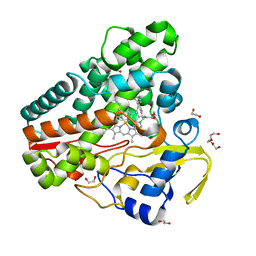 | | The structure of CYP121 in complex with inhibitor L21 | | Descriptor: | 1,2-ETHANEDIOL, 1-[(~{E})-3-[4-(4-fluorophenyl)phenyl]prop-2-enyl]imidazole, Mycocyclosin synthase, ... | | Authors: | Adam, S, Koehnke, J. | | Deposit date: | 2019-11-12 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49986887 Å) | | Cite: | Structure-Activity Relationship and Mode-Of-Action Studies Highlight 1-(4-Biphenylylmethyl)-1H-imidazole-Derived Small Molecules as Potent CYP121 Inhibitors.
Chemmedchem, 16, 2021
|
|
6TEV
 
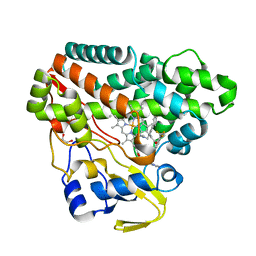 | | The structure of CYP121 in complex with inhibitor L44 | | Descriptor: | 1,2-ETHANEDIOL, 1-[[4-[4-(trifluoromethyl)phenyl]phenyl]methyl]imidazole, Mycocyclosin synthase, ... | | Authors: | Adam, S, Koehnke, J. | | Deposit date: | 2019-11-12 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.70001268 Å) | | Cite: | Structure-Activity Relationship and Mode-Of-Action Studies Highlight 1-(4-Biphenylylmethyl)-1H-imidazole-Derived Small Molecules as Potent CYP121 Inhibitors.
Chemmedchem, 16, 2021
|
|
6TE7
 
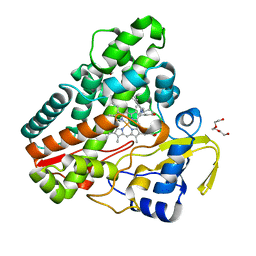 | | The structure of CYP121 in complex with inhibitor S2 | | Descriptor: | 1,2-ETHANEDIOL, 2-chloranyl-4-[4-[(1~{R})-1-imidazol-1-ylprop-2-enyl]phenyl]phenol, Mycocyclosin synthase, ... | | Authors: | Adam, S, Koehnke, J. | | Deposit date: | 2019-11-11 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.50001824 Å) | | Cite: | Structure-Activity Relationship and Mode-Of-Action Studies Highlight 1-(4-Biphenylylmethyl)-1H-imidazole-Derived Small Molecules as Potent CYP121 Inhibitors.
Chemmedchem, 16, 2021
|
|
6ZSV
 
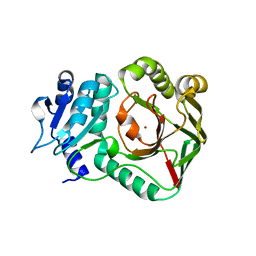 | | Structure of crocagin biosynthetic protein CgnB | | Descriptor: | Uncharacterized protein, ZINC ION | | Authors: | Koehnke, J, Adam, S. | | Deposit date: | 2020-07-16 | | Release date: | 2022-07-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Unusual peptide-binding proteins guide pyrroloindoline alkaloid formation in crocagin biosynthesis.
Nat.Chem., 15, 2023
|
|
1FBZ
 
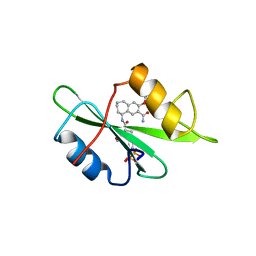 | | Structure-based design of a novel, osteoclast-selective, nonpeptide Src SH2 inhibitor with in vivo anti-resorptive activity | | Descriptor: | PROTO-ONCOGENE TYROSINE-PROTEIN KINASE LCK, {4-[2-ACETYLAMINO-2-(3-CARBAMOYL-2-CYCLOHEXYLMETHOXY-6,7,8,9-TETRAHYDRO-5H-BENZOCYCLOHEPTEN-5YLCARBAMOYL)-ETHYL]-2-PHOSPHONO-PHENYL}-PHOSPHONIC ACID | | Authors: | Shakespeare, W, Yang, M, Bohacek, R, Cerasoli, F, Stebbis, K, Sundaramoorthi, R, Vu, C, Pradeepan, S, Metcalf, C, Haraldson, C, Merry, T, Dalgarno, D, Narula, S, Hatada, M, Lu, X, Van Schravendijk, M.R, Adams, S, Violette, S, Smith, J, Guan, W, Bartlett, C, Herson, J, Iuliucci, J, Weigele, M, Sawyer, T. | | Deposit date: | 2000-07-17 | | Release date: | 2000-08-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based design of an osteoclast-selective, nonpeptide src homology 2 inhibitor with in vivo antiresorptive activity.
Proc.Natl.Acad.Sci.Usa, 97, 2000
|
|
5LHK
 
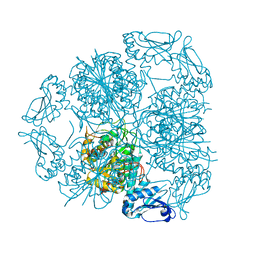 | | Bottromycin maturation enzyme BotP in complex with Mn | | Descriptor: | BICARBONATE ION, Leucine aminopeptidase 2, chloroplastic, ... | | Authors: | Koehnke, J, Adam, S. | | Deposit date: | 2016-07-12 | | Release date: | 2016-10-05 | | Last modified: | 2016-12-14 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure and Substrate Recognition of the Bottromycin Maturation Enzyme BotP.
Chembiochem, 17, 2016
|
|
8A29
 
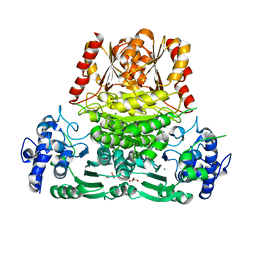 | | Apo 1-deoxy-D-xylulose 5-phosphate synthase from Pseudomonas aeruginosa | | Descriptor: | 1-deoxy-D-xylulose-5-phosphate synthase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hamid, R, Adam, S, Lacour, A, Monjas, L, Hirsch, A. | | Deposit date: | 2022-06-02 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 1-deoxy-D-xylulose-5-phosphate synthase from Pseudomonas aeruginosa and Klebsiella pneumoniae reveals conformational changes upon cofactor binding.
J.Biol.Chem., 299, 2023
|
|
6W8V
 
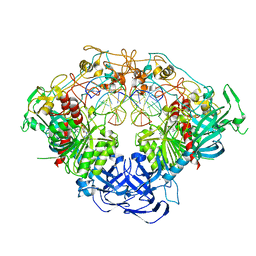 | | Crystal structure of mouse DNMT1 in complex with ACG DNA | | Descriptor: | ACG DNA (5'-D(*AP*CP*TP*TP*AP*(C49)P*GP*GP*AP*AP*GP*G)-3'), ACG DNA (5'-D(*CP*CP*TP*TP*CP*(5CM)P*GP*TP*AP*AP*GP*T)-3'), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Anteneh, H, Song, J. | | Deposit date: | 2020-03-21 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | DNMT1 activity, base flipping mechanism and genome-wide DNA methylation are regulated by the DNA sequence context
Nat Commun, 2020
|
|
7PD7
 
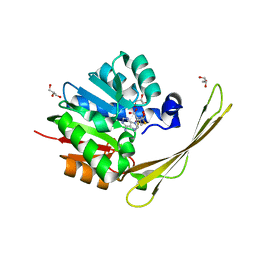 | | Crocagin methyl transferase CgnL | | Descriptor: | GLYCEROL, Methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Zheng, D, Koehnke, J. | | Deposit date: | 2021-08-04 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Unusual peptide-binding proteins guide pyrroloindoline alkaloid formation in crocagin biosynthesis.
Nat.Chem., 15, 2023
|
|
6W8W
 
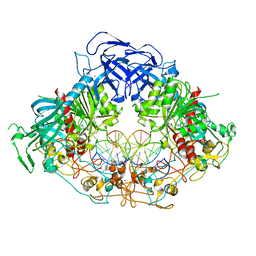 | | Crystal structure of mouse DNMT1 in complex with CCG DNA | | Descriptor: | CCG DNA (5'-D(*AP*CP*TP*TP*AP*(5CM)P*GP*GP*AP*AP*GP*G)-3'), CCG DNA (5'-D(*CP*CP*TP*TP*CP*(C49)P*GP*TP*AP*AP*GP*T)-3'), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Anteneh, H, Song, J. | | Deposit date: | 2020-03-21 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | DNMT1 activity, base flipping mechanism and genome-wide DNA methylation are regulated by the DNA sequence context
Nat Commun, 2020
|
|
5LHJ
 
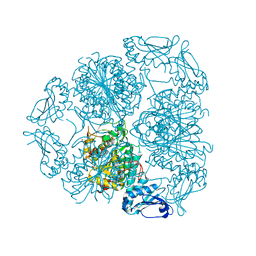 | | Bottromycin maturation enzyme BotP | | Descriptor: | CHLORIDE ION, GLYCEROL, Leucine aminopeptidase 2, ... | | Authors: | Koehnke, J, Mann, G. | | Deposit date: | 2016-07-12 | | Release date: | 2016-10-12 | | Last modified: | 2016-12-14 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure and Substrate Recognition of the Bottromycin Maturation Enzyme BotP.
Chembiochem, 17, 2016
|
|
2BDJ
 
 | | Src kinase in complex with inhibitor AP23464 | | Descriptor: | 3-[2-(2-CYCLOPENTYL-6-{[4-(DIMETHYLPHOSPHORYL)PHENYL]AMINO}-9H-PURIN-9-YL)ETHYL]PHENOL, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Dalgarno, D, Stehle, T, Schelling, P, Narula, S, Sawyer, T. | | Deposit date: | 2005-10-20 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of Src tyrosine kinase inhibition with a new class of potent and selective trisubstituted purine-based compounds.
Chem.Biol.Drug Des., 67, 2006
|
|
2BDF
 
 | | Src kinase in complex with inhibitor AP23451 | | Descriptor: | Proto-oncogene tyrosine-protein kinase Src, {[(4-{[2-(4-AMINOCYCLOHEXYL)-9-ETHYL-9H-PURIN-6-YL]AMINO}PHENYL)(HYDROXY)PHOSPHORYL]METHYL}PHOSPHONIC ACID | | Authors: | Dalgarno, D, Stehle, T, Schelling, P, Sawyer, T, Narula, S. | | Deposit date: | 2005-10-20 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of Src tyrosine kinase inhibition with a new class of potent and selective trisubstituted purine-based compounds.
Chem.Biol.Drug Des., 67, 2006
|
|
6T6H
 
 | | Apo structure of the Bottromycin epimerase BotH | | Descriptor: | BotH, SODIUM ION, SULFATE ION | | Authors: | Koehnke, J, Sikandar, A. | | Deposit date: | 2019-10-18 | | Release date: | 2020-07-15 | | Last modified: | 2020-08-26 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | The bottromycin epimerase BotH defines a group of atypical alpha / beta-hydrolase-fold enzymes.
Nat.Chem.Biol., 16, 2020
|
|
6T6X
 
 | | Structure of the Bottromycin epimerase BotH in complex with substrate | | Descriptor: | (4~{R})-2-[(1~{R})-1-[[(2~{S})-2-[[(2~{S})-3-methyl-2-[[(4~{Z},6~{S},9~{S},12~{S})-2,8,11-tris(oxidanylidene)-6,9-di(propan-2-yl)-1,4,7,10-tetrazabicyclo[10.3.0]pentadec-4-en-5-yl]amino]butanoyl]amino]-3-phenyl-propanoyl]amino]-3-oxidanyl-3-oxidanylidene-propyl]-4,5-dihydro-1,3-thiazole-4-carboxylic acid, BotH | | Authors: | Koehnke, J, Sikandar, A. | | Deposit date: | 2019-10-20 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The bottromycin epimerase BotH defines a group of atypical alpha / beta-hydrolase-fold enzymes.
Nat.Chem.Biol., 16, 2020
|
|
