4W2O
 
 | |
4W2P
 
 | | Anti-Marburgvirus Nucleoprotein Single Domain Antibody C | | Descriptor: | ACETATE ION, Anti-Marburgvirus Nucleoprotein Single Domain Antibody C, SODIUM ION | | Authors: | Taylor, A.B, Garza, J.A. | | Deposit date: | 2017-08-17 | | Release date: | 2017-10-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Unveiling a Drift Resistant Cryptotope withinMarburgvirusNucleoprotein Recognized by Llama Single-Domain Antibodies.
Front Immunol, 8, 2017
|
|
4W2Q
 
 | |
1A8J
 
 | |
5SUO
 
 | |
3MCG
 
 | |
8JSH
 
 | |
8JSG
 
 | |
2MCG
 
 | |
1A56
 
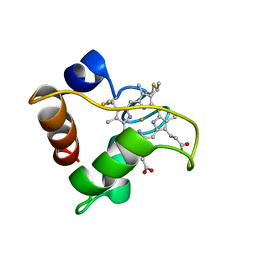 | | PRIMARY SEQUENCE AND SOLUTION CONFORMATION OF FERRICYTOCHROME C-552 FROM NITROSOMONAS EUROPAEA, NMR, MEAN STRUCTURE REFINED WITH EXPLICIT HYDROGEN BOND CONSTRAINTS | | Descriptor: | FERRICYTOCHROME C-552, HEME C | | Authors: | Timkovich, R, Bergmann, D, Arciero, D.M, Hooper, A.B. | | Deposit date: | 1998-02-20 | | Release date: | 1998-10-21 | | Last modified: | 2020-12-16 | | Method: | SOLUTION NMR | | Cite: | Primary sequence and solution conformation of ferrocytochrome c-552 from Nitrosomonas europaea.
Biophys.J., 75, 1998
|
|
1AIH
 
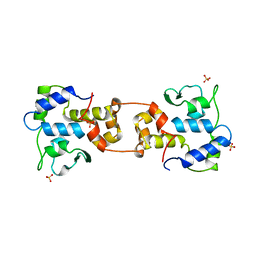 | | CATALYTIC DOMAIN OF BACTERIOPHAGE HP1 INTEGRASE | | Descriptor: | HP1 INTEGRASE, MAGNESIUM ION, SULFATE ION | | Authors: | Hickman, A.B, Waninger, S, Scocca, J.J, Dyda, F. | | Deposit date: | 1997-04-17 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular organization in site-specific recombination: the catalytic domain of bacteriophage HP1 integrase at 2.7 A resolution.
Cell(Cambridge,Mass.), 89, 1997
|
|
1A8C
 
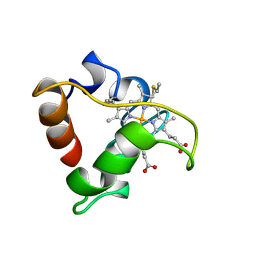 | | PRIMARY SEQUENCE AND SOLUTION CONFORMATION OF FERROCYTOCHROME C-552 FROM NITROSOMONAS EUROPAEA, NMR, MEAN STRUCTURE REFINED WITHOUT HYDROGEN BOND CONSTRAINTS | | Descriptor: | FERROCYTOCHROME C-552, HEME C | | Authors: | Timkovich, R, Bergmann, D, Arciero, D.M, Hooper, A.B. | | Deposit date: | 1998-03-23 | | Release date: | 1998-10-21 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Primary sequence and solution conformation of ferrocytochrome c-552 from Nitrosomonas europaea.
Biophys.J., 75, 1998
|
|
1A8F
 
 | | HUMAN SERUM TRANSFERRIN, RECOMBINANT N-TERMINAL LOBE | | Descriptor: | CARBONATE ION, FE (III) ION, SERUM TRANSFERRIN | | Authors: | Macgillivray, R.T.A, Moore, S.A, Chen, J, Anderson, B.F, Baker, H, Luo, Y, Bewley, M, Smith, C.A, Murphy, M.E.P, Wang, Y, Mason, A.B, Woodworth, R.C, Brayer, G.D, Baker, E.N. | | Deposit date: | 1998-03-25 | | Release date: | 1998-06-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Two high-resolution crystal structures of the recombinant N-lobe of human transferrin reveal a structural change implicated in iron release.
Biochemistry, 37, 1998
|
|
1A8P
 
 | | FERREDOXIN REDUCTASE FROM AZOTOBACTER VINELANDII | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADPH:FERREDOXIN OXIDOREDUCTASE | | Authors: | Prasad, G.S, Kresge, N, Muhlberg, A.B, Shaw, A, Jung, Y.S, Burgess, B.K, Stout, C.D. | | Deposit date: | 1998-03-28 | | Release date: | 1998-06-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of NADPH:ferredoxin reductase from Azotobacter vinelandii.
Protein Sci., 7, 1998
|
|
1A8E
 
 | | HUMAN SERUM TRANSFERRIN, RECOMBINANT N-TERMINAL LOBE | | Descriptor: | CARBONATE ION, FE (III) ION, SERUM TRANSFERRIN | | Authors: | Macgillivray, R.T.A, Moore, S.A, Chen, J, Anderson, B.F, Baker, H, Luo, Y, Bewley, M, Smith, C.A, Murphy, M.E.P, Wang, Y, Mason, A.B, Woodworth, R.C, Brayer, G.D, Baker, E.N. | | Deposit date: | 1998-03-24 | | Release date: | 1998-06-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Two high-resolution crystal structures of the recombinant N-lobe of human transferrin reveal a structural change implicated in iron release.
Biochemistry, 37, 1998
|
|
1AQK
 
 | |
1B2W
 
 | | COMPARISON OF THE THREE-DIMENSIONAL STRUCTURES OF A HUMANIZED AND A CHIMERIC FAB OF AN ANTI-GAMMA-INTERFERON ANTIBODY | | Descriptor: | PROTEIN (ANTIBODY (HEAVY CHAIN)), PROTEIN (ANTIBODY (LIGHT CHAIN)) | | Authors: | Fan, Z, Shan, L, Goldsteen, B.Z, Guddat, L.W, Thakur, A, Landolfi, N.F, Co, M.S, Vasquez, M, Queen, C, Ramsland, P.A, Edmundson, A.B. | | Deposit date: | 1998-12-01 | | Release date: | 1999-05-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Comparison of the three-dimensional structures of a humanized and a chimeric Fab of an anti-gamma-interferon antibody.
J.Mol.Recog., 12, 1999
|
|
1B4J
 
 | | COMPARISON OF THE THREE-DIMENSIONAL STRUCTURES OF A HUMANIZED AND A CHIMERIC FAB OF AN ANTI-GAMMA-INTERFERON ANTIBODY | | Descriptor: | ANTIBODY | | Authors: | Fan, Z, Shan, L, Goldsteen, B.Z, Guddat, L.W, Thakur, A, Landolfi, N.F, Co, M.S, Vasques, M, Queen, C, Ramsland, P.A, Edmundson, A.B. | | Deposit date: | 1998-12-22 | | Release date: | 1999-06-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Comparison of the three-dimensional structures of a humanized and a chimeric Fab of an anti-gamma-interferon antibody.
J.Mol.Recog., 12, 1999
|
|
1BIS
 
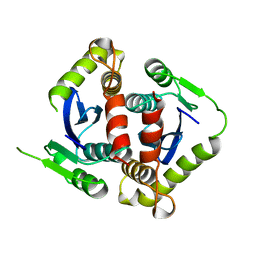 | | HIV-1 INTEGRASE CORE DOMAIN | | Descriptor: | HIV-1 INTEGRASE | | Authors: | Goldgur, Y, Dyda, F, Hickman, A.B, Jenkins, T.M, Craigie, R, Davies, D.R. | | Deposit date: | 1998-06-19 | | Release date: | 1998-08-19 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1BIU
 
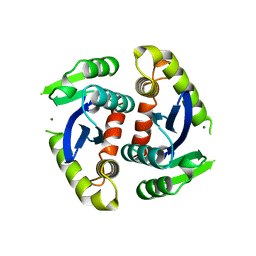 | | HIV-1 INTEGRASE CORE DOMAIN COMPLEXED WITH MG++ | | Descriptor: | HIV-1 INTEGRASE, MAGNESIUM ION | | Authors: | Goldgur, Y, Dyda, F, Hickman, A.B, Jenkins, T.M, Craigie, R, Davies, D.R. | | Deposit date: | 1998-06-19 | | Release date: | 1998-08-19 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1BIZ
 
 | | HIV-1 INTEGRASE CORE DOMAIN | | Descriptor: | CACODYLATE ION, HIV-1 INTEGRASE | | Authors: | Goldgur, Y, Dyda, F, Hickman, A.B, Jenkins, T.M, Craigie, R, Davies, D.R. | | Deposit date: | 1998-06-21 | | Release date: | 1998-08-19 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1C7T
 
 | | BETA-N-ACETYLHEXOSAMINIDASE MUTANT E540D COMPLEXED WITH DI-N ACETYL-D-GLUCOSAMINE (CHITOBIASE) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-N-ACETYLHEXOSAMINIDASE, SULFATE ION | | Authors: | Prag, G, Papanikolau, Y, Tavlas, G, Vorgias, C.E, Petratos, K, Oppenheim, A.B. | | Deposit date: | 2000-03-17 | | Release date: | 2000-09-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of chitobiase mutants complexed with the substrate Di-N-acetyl-d-glucosamine: the catalytic role of the conserved acidic pair, aspartate 539 and glutamate 540.
J.Mol.Biol., 300, 2000
|
|
1CBV
 
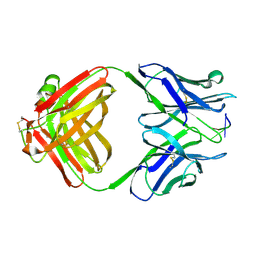 | | AN AUTOANTIBODY TO SINGLE-STRANDED DNA: COMPARISON OF THE THREE-DIMENSIONAL STRUCTURES OF THE UNLIGANDED FAB AND A DEOXYNUCLEOTIDE-FAB COMPLEX | | Descriptor: | DNA (5'-D(*TP*TP*T)-3'), PROTEIN (FAB (BV04-01) AUTOANTIBODY-HEAVY CHAIN), PROTEIN (FAB (BV04-01) AUTOANTIBODY-LIGHT CHAIN) | | Authors: | Herron, J.N, He, X.M, Ballard, D.W, Blier, P.R, Pace, P.E, Bothwell, A.L.M, Voss Junior, E.W, Edmundson, A.B. | | Deposit date: | 1993-03-16 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | An autoantibody to single-stranded DNA: comparison of the three-dimensional structures of the unliganded Fab and a deoxynucleotide-Fab complex.
Proteins, 11, 1991
|
|
7BK5
 
 | | PfCopC mutant - E27A | | Descriptor: | CHLORIDE ION, COPPER (II) ION, PHOSPHATE ION, ... | | Authors: | Muderspach, S.J, Ipsen, J, Rollan, C.H, Bertelsen, A.B, Norholm, M.H.H, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-01-15 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Copper binding and reactivity at the histidine brace motif: insights from mutational analysis of the Pseudomonas fluorescens copper chaperone CopC.
Febs Lett., 595, 2021
|
|
7BK7
 
 | | PfCopC mutant - D83N | | Descriptor: | ACETATE ION, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Muderspach, S.J, Ipsen, J, Rollan, C.H, Bertelsen, A.B, Norholm, M.H.H, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-01-15 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Copper binding and reactivity at the histidine brace motif: insights from mutational analysis of the Pseudomonas fluorescens copper chaperone CopC.
Febs Lett., 595, 2021
|
|
