1CMR
 
 | | NMR SOLUTION STRUCTURE OF A CHIMERIC PROTEIN, DESIGNED BY TRANSFERRING A FUNCTIONAL SNAKE BETA-HAIRPIN INTO A SCORPION ALPHA/BETA SCAFFOLD (PH 3.5, 20C), NMR, 18 STRUCTURES | | Descriptor: | CHARYBDOTOXIN, ALPHA CHIMERA | | Authors: | Zinn-Justin, S, Guenneugues, M, Drakopoulou, E, Gilquin, B, Vita, C, Menez, A. | | Deposit date: | 1996-03-15 | | Release date: | 1996-08-01 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Transfer of a beta-hairpin from the functional site of snake curaremimetic toxins to the alpha/beta scaffold of scorpion toxins: three-dimensional solution structure of the chimeric protein.
Biochemistry, 35, 1996
|
|
1NEA
 
 | | THREE-DIMENSIONAL SOLUTION STRUCTURE OF A CURAREMIMETIC TOXIN FROM NAJA NIGRICOLLIS VENOM: A PROTON NMR AND MOLECULAR MODELING STUDY | | Descriptor: | TOXIN ALPHA | | Authors: | Zinn-Justin, S, Roumestand, C, Gilquin, B, Bontems, F, Menez, A, Toma, F. | | Deposit date: | 1992-09-22 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of a curaremimetic toxin from Naja nigricollis venom: a proton NMR and molecular modeling study.
Biochemistry, 31, 1992
|
|
7ABM
 
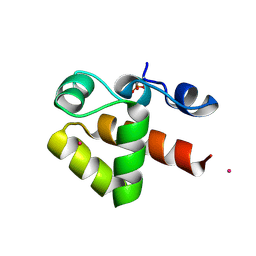 | | X-ray structure of phosphorylated Barrier-to-autointegration factor (BAF) | | Descriptor: | Barrier-to-autointegration factor, CESIUM ION | | Authors: | Zinn-Justin, S, Marcelot, A, Le Du, M.H, Ropars, V. | | Deposit date: | 2020-09-08 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | Di-phosphorylated BAF shows altered structural dynamics and binding to DNA, but interacts with its nuclear envelope partners.
Nucleic Acids Res., 49, 2021
|
|
7NDY
 
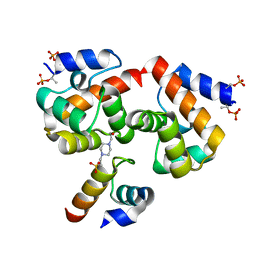 | | Di-phosphorylated Barrier-to-Autointegration Factor (BAF) in complex with LEM domain of Emerin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Barrier-to-autointegration factor, N-terminally processed, ... | | Authors: | Marcelot, A, Le Du, M.H, Hoffmann, G, Zinn-Justin, S. | | Deposit date: | 2021-02-02 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Di-phosphorylated BAF shows altered structural dynamics and binding to DNA, but interacts with its nuclear envelope partners.
Nucleic Acids Res., 49, 2021
|
|
1BF0
 
 | | CALCICLUDINE (CAC) FROM GREEN MAMBA DENDROASPIS ANGUSTICEPS, NMR, 15 STRUCTURES | | Descriptor: | CALCICLUDINE | | Authors: | Gilquin, B, Lecoq, A, Desne, F, Guenneugues, M, Zinn-Justin, S, Menez, A. | | Deposit date: | 1998-05-26 | | Release date: | 1999-01-13 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Conformational and functional variability supported by the BPTI fold: solution structure of the Ca2+ channel blocker calcicludine.
Proteins, 34, 1999
|
|
7OZ0
 
 | |
5LXL
 
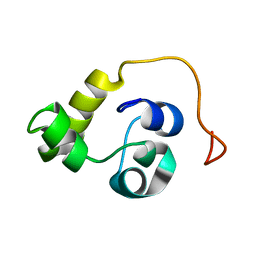 | | NMR structure of the N-terminal domain of the Bacteriophage T5 decoration protein pb10 | | Descriptor: | Decoration protein | | Authors: | Vernhes, E, Gilquin, B, Cuniasse, P, Boulanger, P, Zinn-Justin, S. | | Deposit date: | 2016-09-22 | | Release date: | 2017-04-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | High affinity anchoring of the decoration protein pb10 onto the bacteriophage T5 capsid.
Sci Rep, 7, 2017
|
|
5LXK
 
 | | NMR structure of the C-terminal domain of the Bacteriophage T5 decoration protein pb10. | | Descriptor: | Decoration protein | | Authors: | Vernhes, E, Gilquin, B, Cuniasse, P, Boulanger, P, Zinn-Justin, S. | | Deposit date: | 2016-09-22 | | Release date: | 2017-08-02 | | Last modified: | 2019-05-08 | | Method: | SOLUTION NMR | | Cite: | High affinity anchoring of the decoration protein pb10 onto the bacteriophage T5 capsid.
Sci Rep, 7, 2017
|
|
1CXN
 
 | | REFINED THREE-DIMENSIONAL SOLUTION STRUCTURE OF A SNAKE CARDIOTOXIN: ANALYSIS OF THE SIDE-CHAIN ORGANISATION SUGGESTS THE EXISTENCE OF A POSSIBLE PHOSPHOLIPID BINDING SITE | | Descriptor: | CARDIOTOXIN GAMMA | | Authors: | Gilquin, B, Roumestand, C, Zinn-Justin, S, Menez, A, Toma, F. | | Deposit date: | 1994-07-08 | | Release date: | 1994-12-20 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Refined three-dimensional solution structure of a snake cardiotoxin: analysis of the side-chain organization suggests the existence of a possible phospholipid binding site.
Biopolymers, 33, 1993
|
|
1CXO
 
 | | REFINED THREE-DIMENSIONAL SOLUTION STRUCTURE OF A SNAKE CARDIOTOXIN: ANALYSIS OF THE SIDE-CHAIN ORGANISATION SUGGESTS THE EXISTENCE OF A POSSIBLE PHOSPHOLIPID BINDING SITE | | Descriptor: | CARDIOTOXIN GAMMA | | Authors: | Gilquin, B, Roumestand, C, Zinn-Justin, S, Menez, A, Toma, F. | | Deposit date: | 1994-11-07 | | Release date: | 1994-12-20 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Refined three-dimensional solution structure of a snake cardiotoxin: analysis of the side-chain organization suggests the existence of a possible phospholipid binding site.
Biopolymers, 33, 1993
|
|
7BDX
 
 | | Armadillo domain of HSF2BP in complex with BRCA2 peptide | | Descriptor: | Breast cancer type 2 susceptibility protein, Heat shock factor 2-binding protein, MAGNESIUM ION | | Authors: | Le Du, M.H, Zinn-Justin, S, Ghouil, R, Miron, S, Legrand, P. | | Deposit date: | 2020-12-22 | | Release date: | 2021-07-07 | | Last modified: | 2022-07-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | BRCA2 binding through a cryptic repeated motif to HSF2BP oligomers does not impact meiotic recombination.
Nat Commun, 12, 2021
|
|
6GY2
 
 | |
6YEG
 
 | | Hybrid structure of the SPP1 tail tube by solid-state NMR and cryo EM - Final EM Refinement | | Descriptor: | Tail tube protein gp17.1* | | Authors: | Zinke, M, Sachowsky, K.A.A, Zinn-Justin, S, Ravelli, R, Schroder, G.F, Habeck, M, Lange, A. | | Deposit date: | 2020-03-24 | | Release date: | 2020-10-14 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (4 Å), SOLID-STATE NMR | | Cite: | Architecture of the flexible tail tube of bacteriophage SPP1.
Nat Commun, 11, 2020
|
|
6YQ5
 
 | | Hybrid structure of the SPP1 tail tube by solid-state NMR and cryo EM - NMR Ensemble | | Descriptor: | Tail tube protein gp17.1* | | Authors: | Zinke, M, Sachowsky, K.A.A, Zinn-Justin, S, Ravelli, R, Schroder, G.F, Habeck, M, Lange, A. | | Deposit date: | 2020-04-16 | | Release date: | 2020-10-14 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (4 Å), SOLID-STATE NMR | | Cite: | Architecture of the flexible tail tube of bacteriophage SPP1.
Nat Commun, 11, 2020
|
|
2V1N
 
 | | SOLUTION STRUCTURE OF THE REGION 51-160 OF HUMAN KIN17 REVEALS A WINGED HELIX FOLD | | Descriptor: | PROTEIN KIN HOMOLOG | | Authors: | Carlier, L, Le Maire, A, Gondry, M, Guilhaudis, L, Milazzo, I, Davoust, D, Couprie, J, Gilquin, B, Zinn-Justin, S. | | Deposit date: | 2007-05-27 | | Release date: | 2007-11-27 | | Last modified: | 2020-01-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Region 51-160 of Human Kin17 Reveals an Atypical Winged Helix Domain
Protein Sci., 16, 2007
|
|
8A51
 
 | | Crystal structure of HSF2BP-BRME1 complex | | Descriptor: | 1,2-ETHANEDIOL, Break repair meiotic recombinase recruitment factor 1, CHLORIDE ION, ... | | Authors: | Miron, S, Legrand, P, Ropars, V, Ghouil, R, Zinn-Justin, S. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | BRCA2-HSF2BP oligomeric ring disassembly by BRME1 promotes homologous recombination.
Sci Adv, 9, 2023
|
|
8A50
 
 | | Crystal structure of HSF2BP-ALPHA1 tetramer | | Descriptor: | Heat shock factor 2-binding protein, PHOSPHATE ION | | Authors: | Miron, S, Legrand, P, Ropars, V, Ghouil, R, Zinn-Justin, S. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.484 Å) | | Cite: | BRCA2-HSF2BP oligomeric ring disassembly by BRME1 promotes homologous recombination.
Sci Adv, 9, 2023
|
|
5A20
 
 | | Structure of bacteriophage SPP1 head-to-tail interface filled with DNA and tape measure protein | | Descriptor: | 15 PROTEIN, HEAD COMPLETION PROTEIN GP16, MAJOR TAIL PROTEIN GP17.1, ... | | Authors: | Chaban, Y, Lurz, R, Brasiles, S, Cornilleau, C, Karreman, M, Zinn-Justin, S, Tavares, P, Orlova, E.V. | | Deposit date: | 2015-05-06 | | Release date: | 2015-06-03 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structural Rearrangements in the Phage Head-to-Tail Interface During Assembly and Infection.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5A21
 
 | | Structure of bacteriophage SPP1 head-to-tail interface without DNA and tape measure protein | | Descriptor: | 15 PROTEIN, HEAD COMPLETION PROTEIN GP16, MAJOR TAIL PROTEIN 17.1, ... | | Authors: | Chaban, Y, Lurz, R, Brasiles, S, Cornilleau, C, Karreman, M, Zinn-Justin, S, Tavares, P, Orlova, E.V. | | Deposit date: | 2015-05-06 | | Release date: | 2015-06-03 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Structural Rearrangements in the Phage Head-to-Tail Interface During Assembly and Infection.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7Z21
 
 | | BAF A12T bound to the lamin A/C Ig-fold domain | | Descriptor: | Barrier-to-autointegration factor, N-terminally processed, CHLORIDE ION, ... | | Authors: | Marcelot, A, Legrand, P, Zinn-Justin, S. | | Deposit date: | 2022-02-25 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.629 Å) | | Cite: | The BAF A12T mutation disrupts lamin A/C interaction, impairing robust repair of nuclear envelope ruptures in Nestor-Guillermo progeria syndrome cells.
Nucleic Acids Res., 50, 2022
|
|
2J8A
 
 | | X-ray structure of the N-terminus RRM domain of Set1 | | Descriptor: | HISTONE-LYSINE N-METHYLTRANSFERASE, H3 LYSINE-4 SPECIFIC | | Authors: | Tresaugues, L, Dehe, P.M, Guerois, R, Rodriguez-Gil, A, Varlet, I, Salah, P, Pamblanco, M, Luciano, P, Quevillon-Cheruel, S, Sollier, J, Leulliot, N, Couprie, J, Tordera, V, Zinn-Justin, S, Chavez, S, Van Tilbeurgh, H, Geli, V. | | Deposit date: | 2006-10-24 | | Release date: | 2007-03-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-Ray Structure of the N-Terminus Rrm Domain of Set1
To be Published
|
|
1H9E
 
 | | LEM-LIKE DOMAIN OF HUMAN INNER NUCLEAR MEMBRANE PROTEIN LAP2 | | Descriptor: | LAMINA-ASSOCIATED POLYPEPTIDE 2 | | Authors: | Laguri, C, Gilquin, B, Wolff, N, Romi-Lebrun, R, Courchay, K, Callebaut, I, Worman, H.J, Zinn-Justin, S. | | Deposit date: | 2001-03-08 | | Release date: | 2001-06-17 | | Last modified: | 2017-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of the Lem Motif Common to Three Human Inner Nuclear Membrane Proteins
Structure, 9, 2001
|
|
1H9F
 
 | | LEM DOMAIN OF HUMAN INNER NUCLEAR MEMBRANE PROTEIN LAP2 | | Descriptor: | Lamina-associated polypeptide 2, isoform alpha | | Authors: | Laguri, C, Gilquin, B, Wolff, N, Romi-Lebrun, R, Courchay, K, Callebaut, I, Worman, H.J, Zinn-Justin, S. | | Deposit date: | 2001-03-09 | | Release date: | 2001-06-17 | | Last modified: | 2018-06-13 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of the Lem Motif Common to Three Human Inner Nuclear Membrane Proteins
Structure, 9, 2001
|
|
1QUZ
 
 | | Solution structure of the potassium channel scorpion toxin HSTX1 | | Descriptor: | HSTX1 TOXIN | | Authors: | Savarin, P, Romi-Lebrun, R, Zinn-Justin, S, Lebrun, B, Nakajima, T, Gilquin, B, Menez, A. | | Deposit date: | 1999-07-05 | | Release date: | 2000-07-07 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structural and functional consequences of the presence of a fourth disulfide bridge in the scorpion short toxins: solution structure of the potassium channel inhibitor HsTX1.
Protein Sci., 8, 1999
|
|
1SSF
 
 | | Solution structure of the mouse 53BP1 fragment (residues 1463-1617) | | Descriptor: | Transformation related protein 53 binding protein 1 | | Authors: | Charier, G, Couprie, J, Alpha-Bazin, B, Meyer, V, Quemeneur, E, Guerois, R, Callebaut, I, Gilquin, B, Zinn-Justin, S. | | Deposit date: | 2004-03-24 | | Release date: | 2004-09-14 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The Tudor Tandem of 53BP1; A New Structural Motif Involved in DNA and RG-Rich Peptide Binding
Structure, 12, 2004
|
|
