1OCU
 
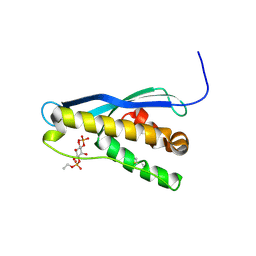 | | Crystal structure of the yeast PX-domain protein Grd19p (sorting nexin 3) complexed to phosphatidylinosytol-3-phosphate. | | Descriptor: | 2-(BUTANOYLOXY)-1-{[(HYDROXY{[2,3,4,6-TETRAHYDROXY-5-(PHOSPHONOOXY)CYCLOHEXYL]OXY}PHOSPHORYL)OXY]METHYL}ETHYL BUTANOATE, SORTING NEXIN | | Authors: | Zhou, C.Z, Li de La Sierra-Gallay, I, Cheruel, S, Collinet, B, Minard, P, Blondeau, K, Henkes, G, Aufrere, R, Leulliot, N, Graille, M, Sorel, I, Savarin, P, de la Torre, F, Poupon, A, Janin, J, van Tilbeurgh, H. | | Deposit date: | 2003-02-10 | | Release date: | 2003-12-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the yeast Phox homology (PX) domain protein Grd19p complexed to phosphatidylinositol-3-phosphate.
J. Biol. Chem., 278, 2003
|
|
1OCS
 
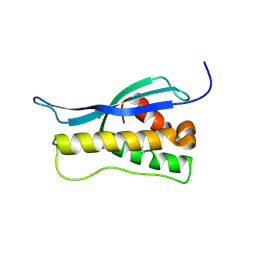 | | Crystal structure of the yeast PX-doamin protein Grd19p (sorting nexin3) complexed to phosphatidylinosytol-3-phosphate. | | Descriptor: | GLYCEROL, SORTING NEXIN GRD19 | | Authors: | Zhou, C.Z, Li De La Sierra-Gallay, I, Cheruel, S, Collinet, B, Minard, P, Blondeau, K, Henkes, G, Aufrere, R, Leulliot, N, Graille, M, Sorel, I, Savarin, P, De La Torre, F, Poupon, A, Janin, J, Van Tilbeurgh, H. | | Deposit date: | 2003-02-10 | | Release date: | 2003-12-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal Structure of the Yeast Phox Homology (Px) Protein Grd19P (Sorting Nexin 3) Complexed to Phosphatidylinositol-3-Phosphate
J.Biol.Chem., 278, 2003
|
|
3F3H
 
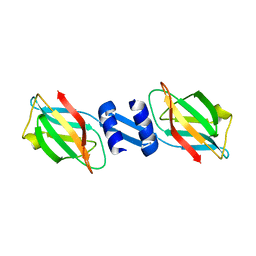 | | Crystal structure and anti-tumor activity of LZ-8 from the fungus Ganoderma lucidium | | Descriptor: | Immunomodulatory protein Ling Zhi-8 | | Authors: | Zhou, C.Z, Huang, L, He, Y.X, Bao, R, Sun, F, Liang, C, Liu, L. | | Deposit date: | 2008-10-30 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of LZ-8 from the medicinal fungus Ganoderma lucidium
Proteins, 75, 2009
|
|
4X36
 
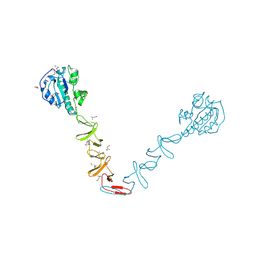 | | Crystal structure of the autolysin LytA from Streptococcus pneumoniae TIGR4 | | Descriptor: | Autolysin, CHOLINE ION, GLYCEROL, ... | | Authors: | Cheng, W, Li, Q, Zhou, C.Z, Chen, Y.X. | | Deposit date: | 2014-11-28 | | Release date: | 2015-05-27 | | Last modified: | 2015-06-24 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Full-length structure of the major autolysin LytA.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8KEF
 
 | |
8KEG
 
 | |
4R78
 
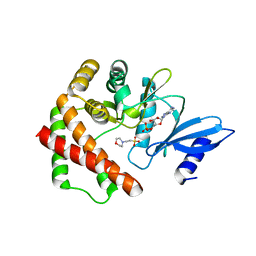 | | Crystal structure of LicA in complex with AMP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE MONOPHOSPHATE, Choline kinase | | Authors: | Wang, L, Jiang, Y.L, Zhou, C.Z, Chen, Y.X. | | Deposit date: | 2014-08-27 | | Release date: | 2015-08-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and enzymatic characterization of the choline kinase LicA from Streptococcus pneumoniae
Plos One, 10, 2015
|
|
4R77
 
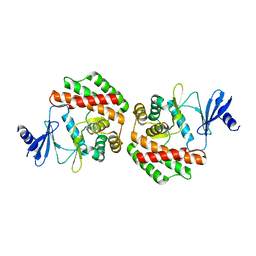 | |
8WXB
 
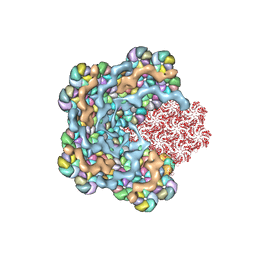 | |
7FDV
 
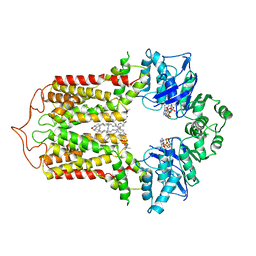 | | Cryo-EM structure of the human cholesterol transporter ABCG1 in complex with cholesterol | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family G member 1, CHOLESTEROL, ... | | Authors: | Xu, D, Li, Y.Y, Yang, F.R, Sun, C.R, Pan, J.H, Wang, L, Chen, Z.P, Fang, S.C, Yao, X.B, Hou, W.T, Zhou, C.Z, Chen, Y. | | Deposit date: | 2021-07-18 | | Release date: | 2022-06-22 | | Last modified: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structure and transport mechanism of the human cholesterol transporter ABCG1.
Cell Rep, 38, 2022
|
|
8HDW
 
 | | Cyanophage Pam3 Sheath-tube | | Descriptor: | Pam3 sheath protein, pam3 tube | | Authors: | Yang, F, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2022-11-06 | | Release date: | 2023-01-18 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Fine structure and assembly pattern of a minimal myophage Pam3.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8HDT
 
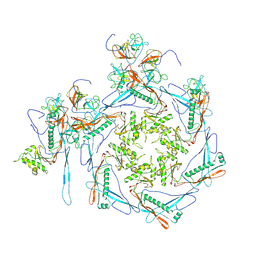 | |
8HDS
 
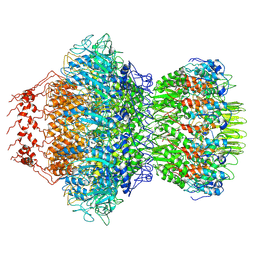 | | Cyanophage Pam3 portal-adaptor | | Descriptor: | Pam3 adaptor protein, Pam3 portal protein | | Authors: | Yang, F, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2022-11-06 | | Release date: | 2023-01-18 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Fine structure and assembly pattern of a minimal myophage Pam3.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8HDR
 
 | | Cyanophage Pam3 neck | | Descriptor: | Pam3 adaptor protein, Pam3 connector protein, Pam3 sheath protein, ... | | Authors: | Yang, F, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2022-11-06 | | Release date: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Fine structure and assembly pattern of a minimal myophage Pam3.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
2HQM
 
 | |
2M80
 
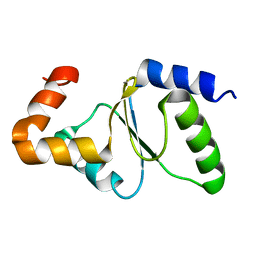 | | Solution structure of yeast dithiol glutaredoxin Grx8 | | Descriptor: | Glutaredoxin-8 | | Authors: | Tang, Y, Zhang, J, Yu, J, Wu, J, Zhou, C.Z, Shi, Y. | | Deposit date: | 2013-05-02 | | Release date: | 2014-05-07 | | Method: | SOLUTION NMR | | Cite: | Structure-guided activity enhancement and catalytic mechanism of yeast grx8
Biochemistry, 53, 2014
|
|
7F38
 
 | |
2P4Q
 
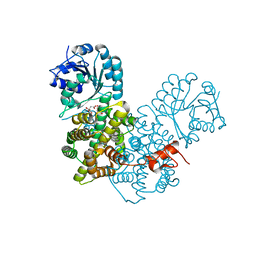 | | Crystal Structure Analysis of Gnd1 in Saccharomyces cerevisiae | | Descriptor: | 6-phosphogluconate dehydrogenase, decarboxylating 1, CITRATE ANION | | Authors: | He, W, Wang, Y, Liu, W, Zhou, C.Z. | | Deposit date: | 2007-03-12 | | Release date: | 2007-07-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structure of Saccharomyces cerevisiae 6-phosphogluconate dehydrogenase Gnd1
Bmc Struct.Biol., 7, 2007
|
|
2QJL
 
 | | Crystal structure of Urm1 | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Ubiquitin-related modifier 1 | | Authors: | Yu, J, Zhou, C.Z. | | Deposit date: | 2007-07-07 | | Release date: | 2007-07-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystal structure of the dimeric Urm1 from the yeast Saccharomyces cerevisiae.
Proteins, 71, 2008
|
|
3FKY
 
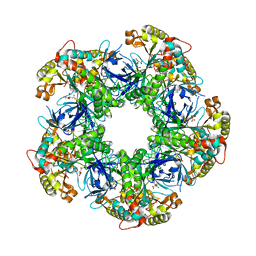 | | Crystal structure of the glutamine synthetase Gln1deltaN18 from the yeast Saccharomyces cerevisiae | | Descriptor: | CITRATE ANION, Glutamine synthetase | | Authors: | He, Y.X, Gui, L, Liu, Y.Z, Du, Y, Zhou, Y.Y, Li, P, Zhou, C.Z. | | Deposit date: | 2008-12-18 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of Saccharomyces cerevisiae glutamine synthetase Gln1 suggests a nanotube-like supramolecular assembly
Proteins, 76, 2009
|
|
4DSR
 
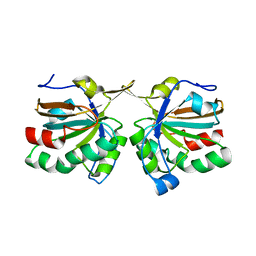 | | Crystal structure of peroxiredoxin Ahp1 from Saccharomyces cerevisiae in reduced form | | Descriptor: | Peroxiredoxin type-2 | | Authors: | Lian, F.M, Yu, J, Ma, X.X, Yu, X.J, Chen, Y, Zhou, C.Z. | | Deposit date: | 2012-02-19 | | Release date: | 2012-04-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structural Snapshots of Yeast Alkyl Hydroperoxide Reductase Ahp1 Peroxiredoxin Reveal a Novel Two-cysteine Mechanism of Electron Transfer to Eliminate Reactive Oxygen Species.
J.Biol.Chem., 287, 2012
|
|
4DSQ
 
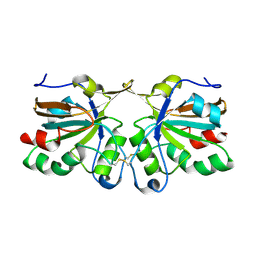 | | Crystal structure of peroxiredoxin Ahp1 from Saccharomyces cerevisiae in oxidized form | | Descriptor: | Peroxiredoxin type-2 | | Authors: | Lian, F.M, Yu, J, Ma, X.X, Yu, X.J, Chen, Y, Zhou, C.Z. | | Deposit date: | 2012-02-19 | | Release date: | 2012-04-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Snapshots of Yeast Alkyl Hydroperoxide Reductase Ahp1 Peroxiredoxin Reveal a Novel Two-cysteine Mechanism of Electron Transfer to Eliminate Reactive Oxygen Species
J.Biol.Chem., 287, 2012
|
|
4E0H
 
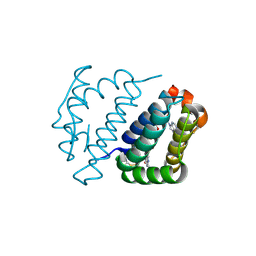 | | Crystal structure of FAD binding domain of Erv1 from Saccharomyces cerevisiae | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Mitochondrial FAD-linked sulfhydryl oxidase ERV1 | | Authors: | Guo, P.C, Ma, J.D, Jiang, Y.L, Wang, S.J, Hu, T.T, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2012-03-04 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of yeast sulfhydryl oxidase erv1 reveals electron transfer of the disulfide relay system in the mitochondrial intermembrane space
J.Biol.Chem., 287, 2012
|
|
4E0I
 
 | | Crystal structure of the C30S/C133S mutant of Erv1 from Saccharomyces cerevisiae | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Mitochondrial FAD-linked sulfhydryl oxidase ERV1 | | Authors: | Guo, P.C, Ma, J.D, Jiang, Y.L, Wang, S.J, Hu, T.T, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2012-03-04 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of yeast sulfhydryl oxidase erv1 reveals electron transfer of the disulfide relay system in the mitochondrial intermembrane space
J.Biol.Chem., 287, 2012
|
|
4DSS
 
 | | Crystal structure of peroxiredoxin Ahp1 from Saccharomyces cerevisiae in complex with thioredoxin Trx2 | | Descriptor: | Peroxiredoxin type-2, Thioredoxin-2 | | Authors: | Lian, F.M, Yu, J, Ma, X.X, Yu, X.J, Chen, Y, Zhou, C.Z. | | Deposit date: | 2012-02-19 | | Release date: | 2012-04-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Snapshots of Yeast Alkyl Hydroperoxide Reductase Ahp1 Peroxiredoxin Reveal a Novel Two-cysteine Mechanism of Electron Transfer to Eliminate Reactive Oxygen Species.
J.Biol.Chem., 287, 2012
|
|
