1CNR
 
 | |
1AB1
 
 | | SI FORM CRAMBIN | | Descriptor: | CRAMBIN (SER22/ILE25), ETHANOL | | Authors: | Teeter, M.M, Yamano, A. | | Deposit date: | 1997-01-31 | | Release date: | 1997-08-12 | | Last modified: | 2016-10-19 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Crystal structure of Ser-22/Ile-25 form crambin confirms solvent, side chain substate correlations.
J.Biol.Chem., 272, 1997
|
|
6KO7
 
 | | Crystal structure of the Ethidium bound RamR determined with XtaLAB Synergy | | Descriptor: | ETHIDIUM, Putative regulatory protein, SULFATE ION | | Authors: | Matsumoto, T, Nakashima, R, Yamano, A, Nishino, K. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Development of a structure determination method using a multidrug-resistance regulator protein as a framework.
Biochem.Biophys.Res.Commun., 518, 2019
|
|
6KO9
 
 | | Crystal structure of the Gefitinib Intermediate 1 bound RamR determined with XtaLAB Synergy | | Descriptor: | 4-[(3-chloranyl-4-fluoranyl-phenyl)amino]-7-methoxy-quinazolin-6-ol, Putative regulatory protein, SULFATE ION | | Authors: | Matsumoto, T, Nakashima, R, Yamano, A, Nishino, K. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Development of a structure determination method using a multidrug-resistance regulator protein as a framework.
Biochem.Biophys.Res.Commun., 518, 2019
|
|
6KO8
 
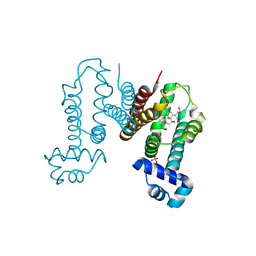 | | Crystal structure of the Cholic acid bound RamR determined with XtaLAB Synergy | | Descriptor: | CHOLIC ACID, Putative regulatory protein, SULFATE ION | | Authors: | Matsumoto, T, Nakashima, R, Yamano, A, Nishino, K. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Development of a structure determination method using a multidrug-resistance regulator protein as a framework.
Biochem.Biophys.Res.Commun., 518, 2019
|
|
5B2H
 
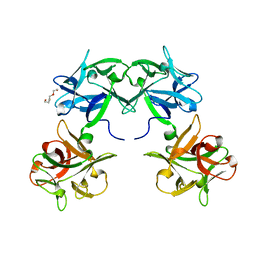 | | Crystal structure of HA33 from Clostridium botulinum serotype C strain Yoichi | | Descriptor: | HA-33, TRIETHYLENE GLYCOL | | Authors: | Akiyama, T, Hayashi, S, Matsumoto, T, Hasegawa, K, Yamano, A, Suzuki, T, Niwa, K, Watanabe, T, Sagane, Y, Yajima, S. | | Deposit date: | 2016-01-15 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational divergence in the HA-33/HA-17 trimer of serotype C and D botulinum toxin complex
Biochem.Biophys.Res.Commun., 476, 2016
|
|
3VUT
 
 | | Crystal structures of non-phosphorylated MAP2K4 | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 4 | | Authors: | Matsumoto, T, Kinoshita, T, Kirii, Y, Tada, T, Yamano, A. | | Deposit date: | 2012-07-05 | | Release date: | 2012-09-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal and solution structures disclose a putative transient state of mitogen-activated protein kinase kinase 4
Biochem.Biophys.Res.Commun., 425, 2012
|
|
3VUO
 
 | | Crystal structure of nontoxic nonhemagglutinin subcomponent (NTNHA) from clostridium botulinum serotype D strain 4947 | | Descriptor: | NTNHA | | Authors: | Sagane, Y, Miyashita, S.-I, Miyata, K, Matsumoto, T, Inui, K, Hayashi, S, Suzuki, T, Hasegawa, K, Yajima, S, Yamano, A, Niwa, K, Watanabe, T. | | Deposit date: | 2012-07-03 | | Release date: | 2012-09-19 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Small-angle X-ray scattering reveals structural dynamics of the botulinum neurotoxin associating protein, nontoxic nonhemagglutinin
Biochem.Biophys.Res.Commun., 425, 2012
|
|
3WL2
 
 | | Monoclinic Lysozyme at 0.96 A resolution | | Descriptor: | 1,2-ETHANEDIOL, Lysozyme C, NITRATE ION, ... | | Authors: | Matsumoto, T, Yamano, A, Hasegawa, T, Maeyama, M. | | Deposit date: | 2013-11-06 | | Release date: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Evaluation of Rigaku XtaLAB P200
To be Published
|
|
3WCQ
 
 | | Crystal structure analysis of Cyanidioschyzon melorae ferredoxin D58N mutant | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Ferredoxin | | Authors: | Ueno, Y, Matsumoto, T, Yamano, A, Imai, T, Morimoto, Y. | | Deposit date: | 2013-05-31 | | Release date: | 2013-08-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Increasing the electron-transfer ability of Cyanidioschyzon merolae ferredoxin by a one-point mutation - A high resolution and Fe-SAD phasing crystal structure analysis of the Asp58Asn mutant
Biochem.Biophys.Res.Commun., 436, 2013
|
|
1IZ9
 
 | | Crystal Structure of Malate Dehydrogenase from Thermus thermophilus HB8 | | Descriptor: | MALATE DEHYDROGENASE | | Authors: | Hirose, R, Hasegawa, T, Yamano, A, Kuramitsu, S, Hamada, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-10-01 | | Release date: | 2002-10-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Malate Dehydrogenase from Thermus themrophilus HB8
To be published
|
|
1JXW
 
 | | CRAMBIN MIXED SEQUENCE FORM AT 180 K. PROTEIN/WATER SUBSTATES | | Descriptor: | Crambin, ETHANOL | | Authors: | Teeter, M.M, Yamano, A, Stec, B, Mohanty, U. | | Deposit date: | 2001-09-10 | | Release date: | 2001-10-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | On the nature of a glassy state of matter in a hydrated protein: Relation to protein function.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1JXT
 
 | | CRAMBIN MIXED SEQUENCE FORM AT 160 K. PROTEIN/WATER SUBSTATES | | Descriptor: | Crambin, ETHANOL | | Authors: | Teeter, M.M, Yamano, A, Stec, B, Mohanty, U. | | Deposit date: | 2001-09-08 | | Release date: | 2001-10-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | On the nature of a glassy state of matter in a hydrated protein: Relation to protein function.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1JXY
 
 | | CRAMBIN MIXED SEQUENCE FORM AT 220 K. PROTEIN/WATER SUBSTATES | | Descriptor: | Crambin, ETHANOL | | Authors: | Teeter, M.M, Yamano, A, Stec, B, Mohanty, U. | | Deposit date: | 2001-09-10 | | Release date: | 2001-10-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | On the nature of a glassy state of matter in a hydrated protein: Relation to protein function.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1JXU
 
 | | CRAMBIN MIXED SEQUENCE FORM AT 240 K. PROTEIN/WATER SUBSTATES | | Descriptor: | Crambin, ETHANOL | | Authors: | Teeter, M.M, Yamano, A, Stec, B, Mohanty, U. | | Deposit date: | 2001-09-09 | | Release date: | 2001-10-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | On the nature of a glassy state of matter in a hydrated protein: Relation to protein function.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1JXX
 
 | | CRAMBIN MIXED SEQUENCE FORM AT 200 K. PROTEIN/WATER SUBSTATES | | Descriptor: | Crambin, ETHANOL | | Authors: | Teeter, M.M, Yamano, A, Stec, B, Mohanty, U. | | Deposit date: | 2001-09-10 | | Release date: | 2001-10-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | On the nature of a glassy state of matter in a hydrated protein: Relation to protein function.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1KRN
 
 | | STRUCTURE OF KRINGLE 4 AT 4C TEMPERATURE AND 1.67 ANGSTROMS RESOLUTION | | Descriptor: | PLASMINOGEN, SULFATE ION | | Authors: | Stec, B, Teeter, M.M, Whitlow, M, Yamano, A. | | Deposit date: | 1995-06-21 | | Release date: | 1997-01-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure of human plasminogen kringle 4 at 1.68 a and 277 K. A possible structural role of disordered residues.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
2D98
 
 | | Structure of VIL (extra KI/I2 added)-xylanase | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Miyatake, H, Hasegawa, T, Yamano, A. | | Deposit date: | 2005-12-09 | | Release date: | 2006-07-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | New methods to prepare iodinated derivatives by vaporizing iodine labelling (VIL) and hydrogen peroxide VIL (HYPER-VIL)
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
2D8P
 
 | | Structure of HYPER-VIL-thaumatin | | Descriptor: | IODIDE ION, Thaumatin I | | Authors: | Miyatake, H, Hasegawa, T, Yamano, A. | | Deposit date: | 2005-12-08 | | Release date: | 2006-07-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | New methods to prepare iodinated derivatives by vaporizing iodine labelling (VIL) and hydrogen peroxide VIL (HYPER-VIL)
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
2D8W
 
 | | Structure of HYPER-VIL-trypsin | | Descriptor: | CALCIUM ION, Cationic trypsin, IODIDE ION | | Authors: | Miyatake, H, Hasegawa, T, Yamano, A. | | Deposit date: | 2005-12-08 | | Release date: | 2006-07-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | New methods to prepare iodinated derivatives by vaporizing iodine labelling (VIL) and hydrogen peroxide VIL (HYPER-VIL)
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
2D91
 
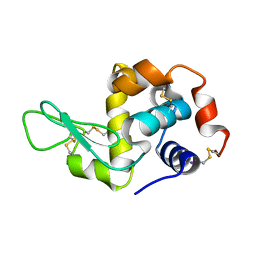 | | Structure of HYPER-VIL-lysozyme | | Descriptor: | IODIDE ION, Lysozyme C | | Authors: | Miyatake, H, Hasegawa, T, Yamano, A. | | Deposit date: | 2005-12-08 | | Release date: | 2006-07-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New methods to prepare iodinated derivatives by vaporizing iodine labelling (VIL) and hydrogen peroxide VIL (HYPER-VIL)
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
2D97
 
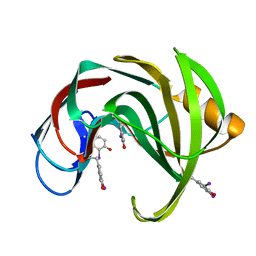 | | Structure of VIL-xylanase | | Descriptor: | Endo-1,4-beta-xylanase 2 | | Authors: | Miyatake, H, Hasegawa, T, Yamano, A. | | Deposit date: | 2005-12-09 | | Release date: | 2006-07-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | New methods to prepare iodinated derivatives by vaporizing iodine labelling (VIL) and hydrogen peroxide VIL (HYPER-VIL)
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
2D8O
 
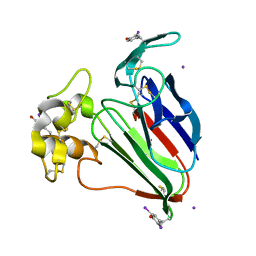 | | Structure of VIL-thaumatin | | Descriptor: | IODIDE ION, Thaumatin I | | Authors: | Miyatake, H, Hasegawa, T, Yamano, A. | | Deposit date: | 2005-12-07 | | Release date: | 2006-07-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | New methods to prepare iodinated derivatives by vaporizing iodine labelling (VIL) and hydrogen peroxide VIL (HYPER-VIL)
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
2DPH
 
 | | Crystal Structure of Formaldehyde dismutase | | Descriptor: | Formaldehyde dismutase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Hasegawa, T, Yamano, A, Yanase, H. | | Deposit date: | 2006-05-11 | | Release date: | 2007-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | The X-ray crystal structure of formaldehyde dismutase at 2.3 A resolution
Acta Crystallogr.,Sect.A, 58, 2002
|
|
2E4M
 
 | | Crystal structure of hemagglutinin subcomponent complex (HA-33/HA-17) from Clostridium botulinum serotype D strain 4947 | | Descriptor: | HA-17, Main hemagglutinin component | | Authors: | Hasegawa, K, Watanabe, T, Suzuki, T, Yamano, A, Niwa, K, Ohyama, T. | | Deposit date: | 2006-12-13 | | Release date: | 2007-06-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Novel Subunit Structure of Clostridium botulinum Serotype D Toxin Complex with Three Extended Arms
J.Biol.Chem., 282, 2007
|
|
