5YJX
 
 | | Structure of the Ndi1 protein from Saccharomyces cerevisiae in complex with myxothiazol. | | Descriptor: | (2Z,6E)-7-{2'-[(2E,4E)-1,6-DIMETHYLHEPTA-2,4-DIENYL]-2,4'-BI-1,3-THIAZOL-4-YL}-3,5-DIMETHOXY-4-METHYLHEPTA-2,6-DIENAMID E, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Yamasita, T, Inaoka, D.K, Shiba, T, Oohashi, T, Iwata, S, Yagi, T, Kosaka, H, Harada, S, Kita, K, Hirano, K. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Ubiquinone binding site of yeast NADH dehydrogenase revealed by structures binding novel competitive- and mixed-type inhibitors
Sci Rep, 8, 2018
|
|
5YJY
 
 | | Structure of the Ndi1 protein from Saccharomyces cerevisiae in complex with AC0-12. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-dodecyl-1-oxidanidyl-quinolin-1-ium-4-ol, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Yamasita, T, Inaoka, D.K, Shiba, T, Oohashi, T, Iwata, S, Yagi, T, Kosaka, H, Harada, S, Kita, K, Hirano, K. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Ubiquinone binding site of yeast NADH dehydrogenase revealed by structures binding novel competitive- and mixed-type inhibitors
Sci Rep, 8, 2018
|
|
5YJW
 
 | | Structure of the Ndi1 protein from Saccharomyces cerevisiae in complex with the competitive inhibitor, stigmatellin. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yamasita, T, Inaoka, D.K, Shiba, T, Oohashi, T, Iwata, S, Yagi, T, Kosaka, H, Harada, S, Kita, K, Hirano, K. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Ubiquinone binding site of yeast NADH dehydrogenase revealed by structures binding novel competitive- and mixed-type inhibitors
Sci Rep, 8, 2018
|
|
6L4P
 
 | | Crystal structure of the complex between the axonemal outer-arm dynein light chain-1 and microtubule binding domain of gamma heavy chain | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Dynein light chain 1, axonemal, ... | | Authors: | Toda, A, Nishikawa, Y, Tanaka, H, Yagi, T, Kurisu, G. | | Deposit date: | 2019-10-19 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | The complex of outer-arm dynein light chain-1 and the microtubule-binding domain of the gamma heavy chain shows how axonemal dynein tunes ciliary beating.
J.Biol.Chem., 295, 2020
|
|
4KEQ
 
 | | Crystal structure of 4-pyridoxolactonase, 5-pyridoxolactone bound | | Descriptor: | 1,2-ETHANEDIOL, 4-pyridoxolactonase, 7-hydroxy-6-methylfuro[3,4-c]pyridin-3(1H)-one, ... | | Authors: | Kobayashi, J, Yoshikane, Y, Baba, S, Mizutani, K, Takahashi, N, Mikami, B, Yagi, T. | | Deposit date: | 2013-04-26 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.279 Å) | | Cite: | Structure of 4-pyridoxolactonase from Mesorhizobium loti.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
4OM8
 
 | | Crystal structure of 5-formly-3-hydroxy-2-methylpyridine 4-carboxylic acid (FHMPC) 5-dehydrogenase, an NAD+ dependent dismutase. | | Descriptor: | 3-hydroxybutyryl-coA dehydrogenase, ACETATE ION, BETA-MERCAPTOETHANOL, ... | | Authors: | Mugo, A.N, Kobayashi, J, Mikami, B, Yagi, T, Ohnishi, K. | | Deposit date: | 2014-01-27 | | Release date: | 2015-01-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of 5-formyl-3-hydroxy-2-methylpyridine 4-carboxylic acid 5-dehydrogenase, an NAD(+)-dependent dismutase from Mesorhizobium loti
Biochem.Biophys.Res.Commun., 456, 2015
|
|
4GAV
 
 | | Structure of the Ndi1 protein from Saccharomyces cerevisiae in complex with quinone | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Rotenone-insensitive NADH-ubiquinone oxidoreductase, UBIQUINONE-2 | | Authors: | Iwata, M, Lee, Y, Yamashita, T, Yagi, T, Iwata, S, Cameron, A.D, Maher, M.J. | | Deposit date: | 2012-07-25 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of the yeast NADH dehydrogenase (Ndi1) reveals overlapping binding sites for water- and lipid-soluble substrates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4G9K
 
 | | Structure of the Ndi1 protein from Saccharomyces cerevisiae | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Rotenone-insensitive NADH-ubiquinone oxidoreductase | | Authors: | Iwata, M, Lee, Y, Yamashita, T, Yagi, T, Iwata, S, Cameron, A.D, Maher, M.J. | | Deposit date: | 2012-07-24 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of the yeast NADH dehydrogenase (Ndi1) reveals overlapping binding sites for water- and lipid-soluble substrates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4GAP
 
 | | Structure of the Ndi1 protein from Saccharomyces cerevisiae in complex with NAD+ | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Rotenone-insensitive NADH-ubiquinone oxidoreductase | | Authors: | Iwata, M, Lee, Y, Yamashita, T, Yagi, T, Iwata, S, Cameron, A.D, Maher, M.J. | | Deposit date: | 2012-07-25 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of the yeast NADH dehydrogenase (Ndi1) reveals overlapping binding sites for water- and lipid-soluble substrates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4HA6
 
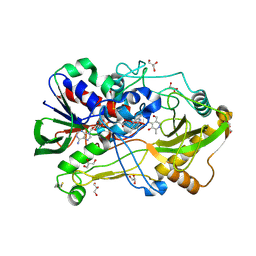 | | Crystal structure of pyridoxine 4-oxidase - pyridoxamine complex | | Descriptor: | 4-(AMINOMETHYL)-5-(HYDROXYMETHYL)-2-METHYLPYRIDIN-3-OL, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Mugo, A.N, Kobayashi, J, Mikami, B, Yagi, T. | | Deposit date: | 2012-09-25 | | Release date: | 2013-06-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | Crystal structure of pyridoxine 4-oxidase from Mesorhizobium loti.
Biochim.Biophys.Acta, 1834, 2013
|
|
2Z9W
 
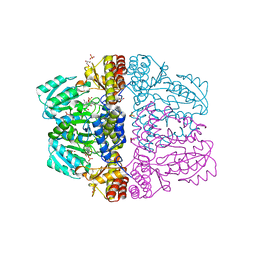 | | Crystal structure of pyridoxamine-pyruvate aminotransferase complexed with pyridoxal | | Descriptor: | 3-HYDROXY-5-(HYDROXYMETHYL)-2-METHYLISONICOTINALDEHYDE, Aspartate aminotransferase, GLYCEROL, ... | | Authors: | Yoshikane, Y, Yokochi, N, Yamasaki, M, Mizutani, K, Ohnishi, K, Mikami, B, Hayashi, H, Yagi, T. | | Deposit date: | 2007-09-26 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of pyridoxamine-pyruvate aminotransferase from Mesorhizobium loti MAFF303099
J.Biol.Chem., 283, 2008
|
|
2Z9X
 
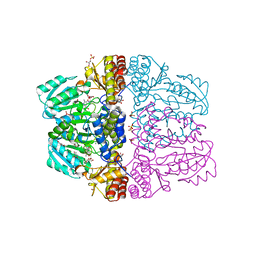 | | Crystal structure of pyridoxamine-pyruvate aminotransferase complexed with pyridoxyl-L-alanine | | Descriptor: | 3-HYDROXY-5-(HYDROXYMETHYL)-2-METHYLISONICOTINALDEHYDE, ALANINE, Aspartate aminotransferase, ... | | Authors: | Yoshikane, Y, Yokochi, N, Yamasaki, M, Mizutani, K, Ohnishi, K, Mikami, B, Hayashi, H, Yagi, T. | | Deposit date: | 2007-09-26 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of pyridoxamine-pyruvate aminotransferase from Mesorhizobium loti MAFF303099
J.Biol.Chem., 283, 2008
|
|
2Z9U
 
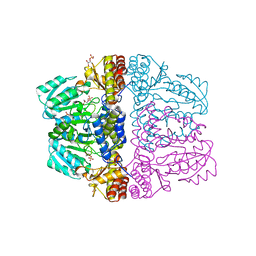 | | Crystal structure of pyridoxamine-pyruvate aminotransferase from Mesorhizobium loti at 2.0 A resolution | | Descriptor: | Aspartate aminotransferase, GLYCEROL, SULFATE ION | | Authors: | Yoshikane, Y, Yokochi, N, Yamasaki, M, Mizutani, K, Ohnishi, K, Mikami, B, Hayashi, H, Yagi, T. | | Deposit date: | 2007-09-26 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of pyridoxamine-pyruvate aminotransferase from Mesorhizobium loti MAFF303099
J.Biol.Chem., 283, 2008
|
|
2Z9V
 
 | | Crystal structure of pyridoxamine-pyruvate aminotransferase complexed with pyridoxamine | | Descriptor: | 4-(AMINOMETHYL)-5-(HYDROXYMETHYL)-2-METHYLPYRIDIN-3-OL, Aspartate aminotransferase, GLYCEROL, ... | | Authors: | Yoshikane, Y, Yokochi, N, Yamasaki, M, Mizutani, K, Ohnishi, K, Mikami, B, Hayashi, H, Yagi, T. | | Deposit date: | 2007-09-26 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of pyridoxamine-pyruvate aminotransferase from Mesorhizobium loti MAFF303099
J.Biol.Chem., 283, 2008
|
|
1WUZ
 
 | | Structure of EC1 domain of CNR | | Descriptor: | Pcdha4 protein | | Authors: | Morishita, H, Umitsu, M, Yamaguchi, T, Murata, Y, Shibata, N, Udaka, K, Higuchi, Y, Akutsu, H, Yagi, T, Ikegami, T. | | Deposit date: | 2004-12-09 | | Release date: | 2005-12-13 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structural diversity of the first cadherin domains revealed by the structure of CNR/Protocadherin alpha
To be Published
|
|
4GF7
 
 | | Crystal structure of 2-Methyl-3-hydroxypyridine-5-carboxylic acid oxygenase (MHPCO), unliganded form | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kobayashi, J, Yoshida, H, Mikami, B, Hayashi, H, Kamitori, S, Sawa, Y, Yagi, T. | | Deposit date: | 2012-08-03 | | Release date: | 2013-08-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase
To be Published
|
|
4H2P
 
 | | Tetrameric form of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase (MHPCO) | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, BETA-MERCAPTOETHANOL, ... | | Authors: | Kobayashi, J, Yoshida, H, Mikami, B, Hayashi, H, Kamitori, S, Yagi, T. | | Deposit date: | 2012-09-13 | | Release date: | 2013-09-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase
To be Published
|
|
4H2N
 
 | | Crystal structure of MHPCO, Y270F mutant | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, BETA-MERCAPTOETHANOL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kobayashi, J, Yoshida, H, Mikami, B, Hayashi, H, Kamitori, S, Yagi, T. | | Deposit date: | 2012-09-12 | | Release date: | 2013-09-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase
To be Published
|
|
4H2Q
 
 | | structure of MHPCO-5HN complex | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, 5-hydroxypyridine-3-carboxylic acid, BETA-MERCAPTOETHANOL, ... | | Authors: | Kobayashi, J, Yoshida, H, Mikami, B, Hayashi, H, Kamitori, S, Yagi, T. | | Deposit date: | 2012-09-13 | | Release date: | 2013-09-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Crystal structure of 2-Methyl-3-hydroxypyridiine-5-carboxylic acid oxygenase
To be Published
|
|
4H2R
 
 | | Structure of MHPCO Y270F mutant, 5-hydroxynicotinic acid complex | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, 5-hydroxypyridine-3-carboxylic acid, BETA-MERCAPTOETHANOL, ... | | Authors: | Kobayashi, J, Yoshida, H, Mikami, B, Hayashi, H, Kamitori, S, Yagi, T. | | Deposit date: | 2012-09-13 | | Release date: | 2013-09-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.473 Å) | | Cite: | Crystal structure of 2-Methyl-3-hydroxypyridine-5-carboxylic acid oxygenase
To be Published
|
|
4JY2
 
 | | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, native and unliganded form | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, BETA-MERCAPTOETHANOL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kobayashi, J, Yoshida, H, Kamitori, S, Hayashi, H, Mizutani, K, Takahashi, N, Mikami, B, Yagi, T. | | Deposit date: | 2013-03-28 | | Release date: | 2014-04-02 | | Method: | X-RAY DIFFRACTION (1.935 Å) | | Cite: | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase
To be Published
|
|
4JY3
 
 | | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, 5-pyridoxic acid bound form | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, 5-hydroxy-4-(hydroxymethyl)-6-methylpyridine-3-carboxylic acid, ... | | Authors: | Kobayashi, J, Yoshida, H, Kamitori, S, Hayashi, H, Mizutani, K, Takahashi, N, Mikami, B, Yagi, T. | | Deposit date: | 2013-03-29 | | Release date: | 2014-04-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase
To be Published
|
|
4KEP
 
 | | Crystal structure of 4-pyridoxolactonase, wild-type | | Descriptor: | 1,2-ETHANEDIOL, 4-pyridoxolactonase, ACETATE ION, ... | | Authors: | Kobayashi, J, Yoshikane, Y, Baba, S, Mizutani, K, Takahashi, N, Mikami, B, Yagi, T. | | Deposit date: | 2013-04-26 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure of 4-pyridoxolactonase from Mesorhizobium loti.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
3ALL
 
 | | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, mutant Y270A | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, BETA-MERCAPTOETHANOL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kobayashi, J, Yoshida, H, Yoshikane, Y, Kamitori, S, Yagi, T. | | Deposit date: | 2010-08-04 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase
To be published
|
|
3ALM
 
 | | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, mutant C294A | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Kobayashi, J, Yoshida, H, Yoshikane, Y, Kamitori, S, Yagi, T. | | Deposit date: | 2010-08-04 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase
To be Published
|
|
