8EPN
 
 | |
8EPQ
 
 | |
8EPP
 
 | |
6MN7
 
 | | Cryo-EM structure of BG505.SOSIP.664 in complex with BF520.1 antigen binding fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BF520.1 Fab variable region, ... | | Authors: | Williams, J.A, Lee, K.K, Overbaugh, J. | | Deposit date: | 2018-10-01 | | Release date: | 2019-03-06 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Kappa chain maturation helps drive rapid development of an infant HIV-1 broadly neutralizing antibody lineage.
Nat Commun, 10, 2019
|
|
1DRS
 
 | | THREE-DIMENSIONAL STRUCTURE OF THE RGD-CONTAINING NEUROTOXIN HOMOLOGUE, DENDROASPIN | | Descriptor: | DENDROASPIN | | Authors: | Sutcliffe, M.J, Jaseja, M, Hyde, E.I, Lu, X, Williams, J.A. | | Deposit date: | 1994-09-29 | | Release date: | 1994-12-20 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the RGD-containing neurotoxin homologue dendroaspin.
Nat.Struct.Biol., 1, 1994
|
|
2HH5
 
 | | Crystal Structure of Cathepsin S in complex with a Zinc mediated non-covalent arylaminoethyl amide | | Descriptor: | CHLORIDE ION, Cathepsin S, N-[(1R)-1-[(BENZYLSULFONYL)METHYL]-2-{[(1S)-1-METHYL-2-{[4-(TRIFLUOROMETHOXY)PHENYL]AMINO}ETHYL]AMINO}-2-OXOETHYL]MORPHOLINE-4-CARBOXAMIDE, ... | | Authors: | Spraggon, G, Hornsby, M, Lesley, S.A, Tully, D.C, Harris, J.L, Karenewsky, D.S. | | Deposit date: | 2006-06-27 | | Release date: | 2006-08-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis and SAR of arylaminoethyl amides as noncovalent inhibitors of cathepsin S: P3 cyclic ethers.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
3E16
 
 | | X-ray structure of human prostasin in complex with Benzoxazole warhead peptidomimic, lysine in P3 | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, Prostasin, ... | | Authors: | Spraggon, G, Hornsby, M, Shipway, A, Harris, J.L, Lesley, S.A. | | Deposit date: | 2008-08-01 | | Release date: | 2008-09-09 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of inhibitors of the channel-activating protease prostasin (CAP1/PRSS8) utilizing structure-based design.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3E0P
 
 | | The X-ray structure of Human Prostasin in complex with a covalent benzoxazole inhibitor | | Descriptor: | GLYCEROL, Prostasin, benzyl [(1R)-1-({(2S,4R)-2-({(1S)-5-amino-1-[(S)-1,3-benzoxazol-2-yl(hydroxy)methyl]pentyl}carbamoyl)-4-[(4-methylbenzyl)oxy]pyrrolidin-1-yl}carbonyl)-3-phenylpropyl]carbamate | | Authors: | Spraggon, G, Hornsby, M, Shipway, A, Harris, J.L, Lesley, S.A. | | Deposit date: | 2008-07-31 | | Release date: | 2008-09-09 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of inhibitors of the channel-activating protease prostasin (CAP1/PRSS8) utilizing structure-based design.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2F1G
 
 | | Cathepsin S in complex with non-covalent 2-(Benzoxazol-2-ylamino)-acetamide | | Descriptor: | Cathepsin S, GLYCEROL, N~2~-1,3-BENZOXAZOL-2-YL-3-CYCLOHEXYL-N-{2-[(4-METHOXYPHENYL)AMINO]ETHYL}-L-ALANINAMIDE | | Authors: | Spraggon, G, Hornsby, M, Lesley, S.A, Tully, D.C, Harris, J.L, Karenewsky, D.S, Kulathila, R, Clark, K. | | Deposit date: | 2005-11-14 | | Release date: | 2006-04-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis and evaluation of arylaminoethyl amides as noncovalent inhibitors of cathepsin S. Part 3: Heterocyclic P3.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2HHN
 
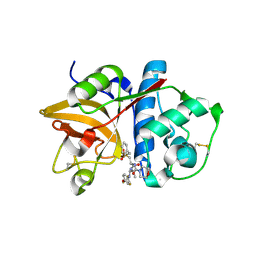 | | Cathepsin S in complex with non covalent arylaminoethyl amide. | | Descriptor: | Cathepsin S, N-[(1R)-1-[(BENZYLSULFONYL)METHYL]-2-{[(1S)-1-METHYL-2-{[4-(TRIFLUOROMETHOXY)PHENYL]AMINO}ETHYL]AMINO}-2-OXOETHYL]MORPHOLINE-4-CARBOXAMIDE, SULFATE ION | | Authors: | Spraggon, G, Hornsby, M, Lesley, S.A, Tully, D.C, Harris, J.L, Karenewsky, D.S, Kulathila, R, Clark, K. | | Deposit date: | 2006-06-28 | | Release date: | 2007-05-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Synthesis and SAR of arylaminoethyl amides as noncovalent inhibitors of cathepsin S: P3 cyclic ethers
Bioorg.Med.Chem.Lett., 16, 2006
|
|
4CLA
 
 | |
