1BLR
 
 | | NMR SOLUTION STRUCTURE OF HUMAN CELLULAR RETINOIC ACID BINDING PROTEIN-TYPE II, 22 STRUCTURES | | Descriptor: | CELLULAR RETINOIC ACID BINDING PROTEIN-TYPE II | | Authors: | Wang, L, Li, Y, Abilddard, F, Yan, H, Markely, J. | | Deposit date: | 1998-07-20 | | Release date: | 1999-01-13 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of type II human cellular retinoic acid binding protein: implications for ligand binding.
Biochemistry, 37, 1998
|
|
4Y7N
 
 | | The Structure Insight into 5-Carboxycytosine Recognition by RNA Polymerase II during Transcription Elongation. | | Descriptor: | DNA (29-MER), DNA (5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, L, Chong, J, Wang, D. | | Deposit date: | 2015-02-15 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Molecular basis for 5-carboxycytosine recognition by RNA polymerase II elongation complex.
Nature, 523, 2015
|
|
4Y52
 
 | | Crystal structure of 5-Carboxycytosine Recognition by RNA Polymerase II during Transcription Elongation. | | Descriptor: | DNA (29-MER), DNA (5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, L, Chong, J, Wang, D. | | Deposit date: | 2015-02-11 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Molecular basis for 5-carboxycytosine recognition by RNA polymerase II elongation complex.
Nature, 523, 2015
|
|
4R78
 
 | | Crystal structure of LicA in complex with AMP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE MONOPHOSPHATE, Choline kinase | | Authors: | Wang, L, Jiang, Y.L, Zhou, C.Z, Chen, Y.X. | | Deposit date: | 2014-08-27 | | Release date: | 2015-08-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and enzymatic characterization of the choline kinase LicA from Streptococcus pneumoniae
Plos One, 10, 2015
|
|
4R77
 
 | |
3Q4A
 
 | |
3Q47
 
 | |
3Q49
 
 | |
6VP0
 
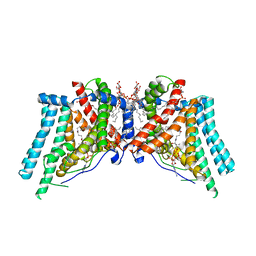 | | Human Diacylglycerol Acyltransferase 1 in complex with oleoyl-CoA | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Diacylglycerol O-acyltransferase 1, Lauryl Maltose Neopentyl Glycol, ... | | Authors: | Wang, L, Qian, H, Han, Y, Nian, Y, Ren, Z, Zhang, H, Hu, L, Prasad, B.V.V, Yan, N, Zhou, M. | | Deposit date: | 2020-02-01 | | Release date: | 2020-05-13 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure and mechanism of human diacylglycerol O-acyltransferase 1.
Nature, 581, 2020
|
|
3TK9
 
 | | Crystal structure of human granzyme H | | Descriptor: | Granzyme H, SULFATE ION | | Authors: | Wang, L, Zhang, K, Wu, L, Tong, L, Sun, F, Fan, Z. | | Deposit date: | 2011-08-25 | | Release date: | 2011-12-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the substrate specificity of human granzyme H: the functional roles of a novel RKR motif
J.Immunol., 188, 2012
|
|
3TJV
 
 | | Crystal structure of human granzyme H with a peptidyl substrate | | Descriptor: | Granzyme H, PTSYAGDDSG, SULFATE ION | | Authors: | Wang, L, Zhang, K, Wu, L, Tong, L, Sun, F, Fan, Z. | | Deposit date: | 2011-08-25 | | Release date: | 2011-12-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the substrate specificity of human granzyme H: the functional roles of a novel RKR motif
J.Immunol., 188, 2012
|
|
3TJU
 
 | | Crystal structure of human granzyme H with an inhibitor | | Descriptor: | Ac-PTSY-CMK inhibitor, Granzyme H, SULFATE ION | | Authors: | Wang, L, Zhang, K, Wu, L, Tong, L, Sun, F, Fan, Z. | | Deposit date: | 2011-08-25 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the substrate specificity of human granzyme H: the functional roles of a novel RKR motif
J.Immunol., 188, 2012
|
|
6PE0
 
 | | Msp1 (E214Q)-substrate complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Membrane-spanning ATPase-like protein, ... | | Authors: | Wang, L, Myasnikov, A, Pan, X, Walter, P. | | Deposit date: | 2019-06-19 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the AAA protein Msp1 reveals mechanism of mislocalized membrane protein extraction.
Elife, 9, 2020
|
|
6PDY
 
 | | Msp1-substrate complex in open conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Wang, L, Myasnikov, A, Pan, X, Walter, P. | | Deposit date: | 2019-06-19 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of the AAA protein Msp1 reveals mechanism of mislocalized membrane protein extraction.
Elife, 9, 2020
|
|
6PDW
 
 | | Msp1-substrate complex in closed conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Wang, L, Myasnikov, A, Pan, X, Walter, P. | | Deposit date: | 2019-06-19 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the AAA protein Msp1 reveals mechanism of mislocalized membrane protein extraction.
Elife, 9, 2020
|
|
6EGA
 
 | | IRAK4 in complex with a type II inhibitor | | Descriptor: | 3-{2-[(cyclopropanecarbonyl)amino][1,3]thiazolo[5,4-b]pyridin-5-yl}-N-{4-[(piperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzamide, COBALT (II) ION, Interleukin-1 receptor-associated kinase 4, ... | | Authors: | Wang, L, Wu, H. | | Deposit date: | 2018-08-19 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.512 Å) | | Cite: | Conformational flexibility and inhibitor binding to unphosphorylated interleukin-1 receptor-associated kinase 4 (IRAK4).
J.Biol.Chem., 294, 2019
|
|
6EG9
 
 | | IRAK4 in complex with Ponatinib | | Descriptor: | 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, COBALT (II) ION, Interleukin-1 receptor-associated kinase 4, ... | | Authors: | Wang, L, Wu, H. | | Deposit date: | 2018-08-19 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.414 Å) | | Cite: | Conformational flexibility and inhibitor binding to unphosphorylated interleukin-1 receptor-associated kinase 4 (IRAK4).
J.Biol.Chem., 294, 2019
|
|
6FXD
 
 | | Crystal structure of MupZ from Pseudomonas fluorescens | | Descriptor: | MupZ | | Authors: | Wang, L, Parnell, A, Williams, C, Bakar, N.A, van der Kamp, M.W, Simpson, T.J, Race, P.R, Crump, M.P, Willis, C.L. | | Deposit date: | 2018-03-08 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A Rieske oxygenase/epoxide hydrolase-catalysed reaction cascade creates oxygen heterocycles in mupirocin biosynthesis
Nat Catal, 2018
|
|
6JFI
 
 | | The symmetric-reconstructed cryo-EM structure of Zika virus-FabZK2B10 complex | | Descriptor: | FabZK2B10 heavy chain, FabZK2B10 light chain, ZIKV structural protein E, ... | | Authors: | Wang, L, Wang, R.K, Wang, L, Ben, H.J, Yu, L, Gao, F, Shi, X.L, Yin, C.B, Zhang, F.C, Xiang, Y, Zhang, L.Q. | | Deposit date: | 2019-02-08 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Structural Basis for Neutralization and Protection by a Zika Virus-Specific Human Antibody.
Cell Rep, 26, 2019
|
|
6JFH
 
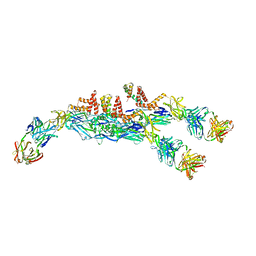 | | The asymmetric-reconstructed cryo-EM structure of Zika virus-FabZK2B10 complex | | Descriptor: | FabZK2B10 heavy chain, FabZK2B10 light chain, ZIKV structural E protein, ... | | Authors: | Wang, L, Wang, R.K, Wang, L, Ben, H.J, Yu, L, Gao, F, Shi, X.L, Yin, C.B, Zhang, F.C, Xiang, Y, Zhang, L.Q. | | Deposit date: | 2019-02-08 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Structural Basis for Neutralization and Protection by a Zika Virus-Specific Human Antibody.
Cell Rep, 26, 2019
|
|
6JEP
 
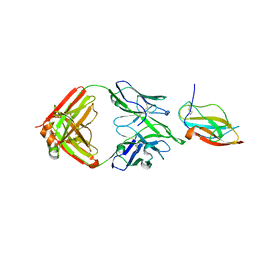 | | Structure of a neutralizing antibody bound to the Zika envelope protein domain III | | Descriptor: | Genome polyprotein, heavy chain of Fab ZK2B10, light chain of Fab ZK2B10 | | Authors: | Wang, L, Wang, R.K, Wang, L, Ben, H.J, Yu, L, Gao, F, Shi, X.L, Yin, C.B, Zhang, F.C, Xiang, Y, Zhang, L.Q. | | Deposit date: | 2019-02-07 | | Release date: | 2019-05-15 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (2.316 Å) | | Cite: | Structural Basis for Neutralization and Protection by a Zika Virus-Specific Human Antibody.
Cell Rep, 26, 2019
|
|
1MLW
 
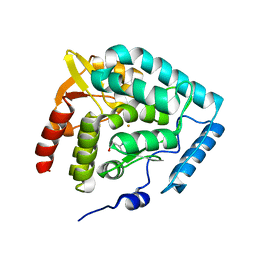 | | Crystal structure of human tryptophan hydroxylase with bound 7,8-dihydro-L-biopterin cofactor and Fe(III) | | Descriptor: | 7,8-DIHYDROBIOPTERIN, FE (III) ION, Tryptophan 5-monooxygenase | | Authors: | Wang, L, Erlandsen, H, Haavik, J, Knappskog, P.M, Stevens, R.C. | | Deposit date: | 2002-08-31 | | Release date: | 2002-12-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Three-dimensional structure of human tryptophan hydroxylase and its implications for the biosynthesis of the neurotransmitters serotonin and melatonin
Biochemistry, 41, 2002
|
|
7LHV
 
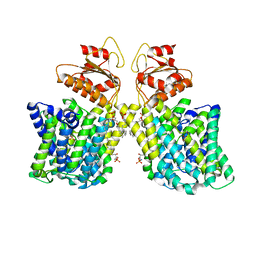 | | Structure of Arabidopsis thaliana sulfate transporter AtSULTR4;1 | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, SULFATE ION, ... | | Authors: | Wang, L, Chen, K, Zhou, M. | | Deposit date: | 2021-01-26 | | Release date: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structure and function of an Arabidopsis thaliana sulfate transporter.
Nat Commun, 12, 2021
|
|
4GA7
 
 | | Crystal structure of human serpinB1 mutant | | Descriptor: | Leukocyte elastase inhibitor | | Authors: | Wang, L, Li, Q, Wu, L, Zhang, K, Tong, L, Sun, F, Fan, Z. | | Deposit date: | 2012-07-25 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification of SERPINB1 as a physiological inhibitor of human granzyme H
J.Immunol., 190, 2013
|
|
4GAW
 
 | | Crystal structure of active human granzyme H | | Descriptor: | CHLORIDE ION, Granzyme H, SULFATE ION | | Authors: | Wang, L, Li, Q, Wu, L, Zhang, K, Tong, L, Sun, F, Fan, Z. | | Deposit date: | 2012-07-25 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Identification of SERPINB1 as a physiological inhibitor of human granzyme H
J.Immunol., 190, 2013
|
|
