7U5C
 
 | | Cryo-EM structure of human CST bound to DNA polymerase alpha-primase in a recruitment state | | Descriptor: | CST complex subunit CTC1, CST complex subunit STN1, CST complex subunit TEN1, ... | | Authors: | Cai, S.W, Zinder, J.C, Svetlov, V, Bush, M.W, Nudler, E, Walz, T, de Lange, T. | | Deposit date: | 2022-03-02 | | Release date: | 2022-05-18 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Cryo-EM structure of the human CST-Pol alpha /primase complex in a recruitment state.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6XAS
 
 | | CryoEM Structure of E. coli Rho-dependent Transcription Pre-termination Complex | | Descriptor: | DNA (29-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Hao, Z.T, Kim, H.K, Walz, T, Nudler, E. | | Deposit date: | 2020-06-04 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Pre-termination Transcription Complex: Structure and Function.
Mol.Cell, 81, 2021
|
|
1FQY
 
 | | STRUCTURE OF AQUAPORIN-1 AT 3.8 A RESOLUTION BY ELECTRON CRYSTALLOGRAPHY | | Descriptor: | AQUAPORIN-1 | | Authors: | Murata, K, Mitsuoka, K, Hirai, T, Walz, T, Agre, P, Heymann, J.B, Engel, A, Fujiyoshi, Y. | | Deposit date: | 2000-09-07 | | Release date: | 2000-10-18 | | Last modified: | 2024-02-07 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.8 Å) | | Cite: | Structural determinants of water permeation through aquaporin-1.
Nature, 407, 2000
|
|
7SD3
 
 | | Cytoplasmic tail deleted HIV-1 Env bound with three 4E10 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4E10 Fab heavy chain, ... | | Authors: | Yang, S, Walz, T. | | Deposit date: | 2021-09-29 | | Release date: | 2022-11-09 | | Last modified: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Dynamic HIV-1 spike motion creates vulnerability for its membrane-bound tripod to antibody attack.
Nat Commun, 13, 2022
|
|
7SC5
 
 | | Cytoplasmic tail deleted HIV Env trimer in nanodisc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Yang, S, Walz, T. | | Deposit date: | 2021-09-27 | | Release date: | 2022-11-09 | | Last modified: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Dynamic HIV-1 spike motion creates vulnerability for its membrane-bound tripod to antibody attack.
Nat Commun, 13, 2022
|
|
1XI5
 
 | | Clathrin D6 coat with auxilin J-domain | | Descriptor: | Auxilin J-domain, Clathrin heavy chain | | Authors: | Fotin, A, Cheng, Y, Grigorieff, N, Walz, T, Harrison, S.C, Kirchhausen, T. | | Deposit date: | 2004-09-21 | | Release date: | 2004-11-02 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structure of an auxilin-bound clathrin coat and its implications for the mechanism of uncoating
Nature, 432, 2004
|
|
1XI4
 
 | | Clathrin D6 Coat | | Descriptor: | Clathrin heavy chain, Clathrin light chain A | | Authors: | Fotin, A, Cheng, Y, Sliz, P, Grigorieff, N, Harrison, S.C, Kirchhausen, T, Walz, T. | | Deposit date: | 2004-09-21 | | Release date: | 2004-11-02 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Molecular model for a complete clathrin lattice from electron cryomicroscopy
Nature, 432, 2004
|
|
3IYV
 
 | | Clathrin D6 coat as full-length Triskelions | | Descriptor: | Clathrin heavy chain, Clathrin light chain A | | Authors: | Johnson, G.T, Fotin, A, Cheng, Y, Sliz, P, Grigorieff, N, Harrison, S.C, Kirchhausen, T, Walz, T. | | Deposit date: | 2010-06-17 | | Release date: | 2010-07-21 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Molecular model for a complete clathrin lattice from electron cryomicroscopy.
Nature, 432, 2004
|
|
6CHS
 
 | | Cdc48-Npl4 complex in the presence of ATP-gamma-S | | Descriptor: | MAGNESIUM ION, Npl4, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Kim, K.H, Bodnar, N.O, Walz, T, Rapoport, T.A. | | Deposit date: | 2018-02-22 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure of the Cdc48 ATPase with its ubiquitin-binding cofactor Ufd1-Npl4.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DNH
 
 | | Cryo-EM structure of human CPSF-160-WDR33-CPSF-30-PAS RNA complex at 3.4 A resolution | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 1, Cleavage and polyadenylation specificity factor subunit 4, RNA (5'-R(P*AP*AP*UP*AP*AP*AP*C)-3'), ... | | Authors: | Sun, Y, Zhang, Y, Hamilton, K, Walz, T, Tong, L. | | Deposit date: | 2018-06-06 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis for the recognition of the human AAUAAA polyadenylation signal.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6OR6
 
 | | Full-length S. pombe Mdn1 in the presence of AMPPNP (tail region) | | Descriptor: | Midasin | | Authors: | Chen, Z, Suzuki, H, Wang, A.C, DiMaio, F, Walz, T, Kapoor, T.M. | | Deposit date: | 2019-04-29 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Structural Insights into Mdn1, an Essential AAA Protein Required for Ribosome Biogenesis.
Cell, 175, 2018
|
|
6ORB
 
 | | Full-length S. pombe Mdn1 in the presence of ATP and Rbin-1 | | Descriptor: | Midasin | | Authors: | Chen, Z, Suzuki, H, Wang, A.C, DiMaio, F, Walz, T, Kapoor, T.M. | | Deposit date: | 2019-04-29 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structural Insights into Mdn1, an Essential AAA Protein Required for Ribosome Biogenesis.
Cell, 175, 2018
|
|
6OR5
 
 | | Full-length S. pombe Mdn1 in the presence of AMPPNP (ring region) | | Descriptor: | Midasin, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Chen, Z, Suzuki, H, Wang, A.C, DiMaio, F, Walz, T, Kapoor, T.M. | | Deposit date: | 2019-04-29 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural Insights into Mdn1, an Essential AAA Protein Required for Ribosome Biogenesis.
Cell, 175, 2018
|
|
2OF5
 
 | | Oligomeric Death Domain complex | | Descriptor: | Death domain-containing protein CRADD, Leucine-rich repeat and death domain-containing protein | | Authors: | Park, H.H, Logette, E, Raunser, S, Cuenin, S, Walz, T, Tschopp, J, Wu, H. | | Deposit date: | 2007-01-02 | | Release date: | 2007-04-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Death domain assembly mechanism revealed by crystal structure of the oligomeric PIDDosome core complex.
Cell(Cambridge,Mass.), 128, 2007
|
|
1SOR
 
 | | Aquaporin-0 membrane junctions reveal the structure of a closed water pore | | Descriptor: | Aquaporin-0 | | Authors: | Gonen, T, Sliz, P, Kistler, J, Cheng, Y, Walz, T. | | Deposit date: | 2004-03-15 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | ELECTRON CRYSTALLOGRAPHY (3 Å) | | Cite: | Aquaporin-0 membrane junctions reveal the structure of a closed water pore
Nature, 429, 2004
|
|
2B6P
 
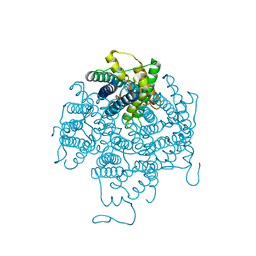 | | X-ray structure of lens Aquaporin-0 (AQP0) (lens MIP) in an open pore state | | Descriptor: | Lens fiber major intrinsic protein | | Authors: | Gonen, T, Cheng, Y, Sliz, P, Hiroaki, Y, Fujiyoshi, Y, Harrison, S.C, Walz, T. | | Deposit date: | 2005-10-03 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Lipid-protein interactions in double-layered two-dimensional AQP0 crystals.
Nature, 438, 2005
|
|
2B6O
 
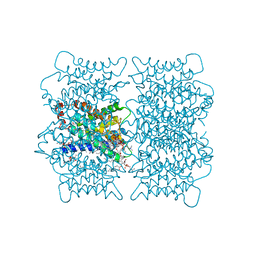 | | Electron crystallographic structure of lens Aquaporin-0 (AQP0) (lens MIP) at 1.9A resolution, in a closed pore state | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Lens fiber major intrinsic protein | | Authors: | Gonen, T, Cheng, Y, Sliz, P, Hiroaki, Y, Fujiyoshi, Y, Harrison, S.C, Walz, T. | | Deposit date: | 2005-10-03 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.9 Å) | | Cite: | Lipid-protein interactions in double-layered two-dimensional AQP0 crystals.
Nature, 438, 2005
|
|
4YCG
 
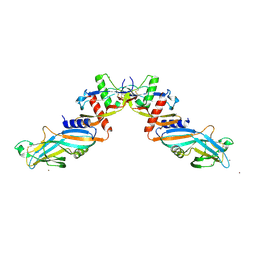 | | Pro-bone morphogenetic protein 9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Bone Morphogenetic Protein 9 Growth Factor Domain, Bone Morphogenetic Protein 9 Prodomain, ... | | Authors: | Mi, L.-Z, Brown, C.T, Gao, Y, Tian, Y, Le, V, Walz, T, Springer, T.A. | | Deposit date: | 2015-02-20 | | Release date: | 2015-03-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of bone morphogenetic protein 9 procomplex.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4YCI
 
 | | non-latent pro-bone morphogenetic protein 9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Bone Morphogenetic Protein 9 Growth Factor Domain, ... | | Authors: | Mi, L.Z, Brown, C.T, Gao, Y, Tian, Y, Le, V, Walz, T, Springer, T.A. | | Deposit date: | 2015-02-20 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure of bone morphogenetic protein 9 procomplex.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5A6E
 
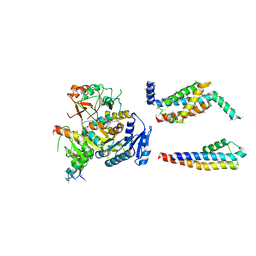 | | Cryo-EM structure of the Slo2.2 Na-activated K channel | | Descriptor: | GATING RING OF POTASSIUM CHANNEL SUBFAMILY T MEMBER 1, PORE DOMAIN OF POTASSIUM CHANNEL SUBFAMILY T MEMBER 1, RCK2 ELABORATION OF POTASSIUM CHANNEL SUBFAMILY T MEMBER 1, ... | | Authors: | Hite, R.K, Yuan, P, Li, Z, Hsuing, Y, Walz, T, MacKinnon, R. | | Deposit date: | 2015-06-25 | | Release date: | 2015-10-14 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-Electron Microscopy Structure of the Slo2.2 Na1-Activated K1 Channel
Nature, 527, 2015
|
|
5A6G
 
 | | Cryo-EM structure of the Slo2.2 Na-activated K channel | | Descriptor: | PORE DOMAIN OF POTASSIUM CHANNEL SUBFAMILY T MEMBER 1, S1-S4 DOMAIN OF POTASSIUM CHANNEL SUBFAMILY T MEMBER 1 | | Authors: | Hite, R.K, Yuan, P, Li, Z, Hsuing, Y, Walz, T, MacKinnon, R. | | Deposit date: | 2015-06-25 | | Release date: | 2015-10-14 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-Electron Microscopy Structure of the Slo2.2 Na1-Activated K1 Channel
Nature, 527, 2015
|
|
5A6F
 
 | | Cryo-EM structure of the Slo2.2 Na-activated K channel | | Descriptor: | GATING RING OF POTASSIUM CHANNEL SUBFAMILY T MEMBER 1, RCK2 ELABORATION OF POTASSIUM CHANNEL SUBFAMILY T MEMBER 1 | | Authors: | Hite, R.K, Yuan, P, Li, Z, Hsuing, Y, Walz, T, MacKinnon, R. | | Deposit date: | 2015-06-25 | | Release date: | 2015-10-14 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-Electron Microscopy Structure of the Slo2.2 Na1-Activated K1 Channel
Nature, 527, 2015
|
|
7LA4
 
 | | Integrin AlphaIIbBeta3-PT25-2 Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Bush, M.W, Walz, T, Coller, B, Filizola, M, Spasic, A, Nesic, D, Li, J. | | Deposit date: | 2021-01-05 | | Release date: | 2022-01-12 | | Last modified: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Electron microscopy shows that binding of monoclonal antibody PT25-2 primes integrin alpha IIb beta 3 for ligand binding.
Blood Adv, 5, 2021
|
|
5TTP
 
 | |
5TV4
 
 | | 3D cryo-EM reconstruction of nucleotide-free MsbA in lipid nanodisc | | Descriptor: | 3-HYDROXY-TETRADECANOIC ACID, L-glycero-alpha-D-manno-heptopyranose-(1-7)-L-glycero-alpha-D-manno-heptopyranose-(1-3)-L-glycero-alpha-D-manno-heptopyranose-(1-5)-[3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)]3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-6)-2-amino-2-deoxy-alpha-D-glucopyranose-(1-6)-2-amino-2-deoxy-alpha-D-glucopyranose, LAURIC ACID, ... | | Authors: | Mi, W, Walz, T, Liao, M. | | Deposit date: | 2016-11-08 | | Release date: | 2017-09-20 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis of MsbA-mediated lipopolysaccharide transport.
Nature, 549, 2017
|
|
