1UMT
 
 | | Stromelysin-1 catalytic domain with hydrophobic inhibitor bound, ph 7.0, 32oc, 20 mm cacl2, 15% acetonitrile; nmr average of 20 structures minimized with restraints | | Descriptor: | CALCIUM ION, N-{(2R)-2-[2-(hydroxyamino)-2-oxoethyl]-4-methylpentanoyl}-L-leucyl-L-phenylalaninamide, STROMELYSIN-1, ... | | Authors: | Van Doren, S.R, Kurochkin, A.V, Hu, W, Zuiderweg, E.R.P. | | Deposit date: | 1995-10-31 | | Release date: | 1996-03-08 | | Last modified: | 2012-12-12 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the catalytic domain of human stromelysin complexed with a hydrophobic inhibitor.
Protein Sci., 4, 1995
|
|
1UMS
 
 | | STROMELYSIN-1 CATALYTIC DOMAIN WITH HYDROPHOBIC INHIBITOR BOUND, PH 7.0, 32OC, 20 MM CACL2, 15% ACETONITRILE; NMR ENSEMBLE OF 20 STRUCTURES | | Descriptor: | CALCIUM ION, N-{(2R)-2-[2-(hydroxyamino)-2-oxoethyl]-4-methylpentanoyl}-L-leucyl-L-phenylalaninamide, STROMELYSIN-1, ... | | Authors: | Van Doren, S.R, Kurochkin, A.V, Hu, W, Zuiderweg, E.R.P. | | Deposit date: | 1995-10-31 | | Release date: | 1996-03-08 | | Last modified: | 2020-09-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the catalytic domain of human stromelysin complexed with a hydrophobic inhibitor.
Protein Sci., 4, 1995
|
|
1D2B
 
 | | THE MMP-INHIBITORY, N-TERMINAL DOMAIN OF HUMAN TISSUE INHIBITOR OF METALLOPROTEINASES-1 (N-TIMP-1), SOLUTION NMR, 29 STRUCTURES | | Descriptor: | Metalloproteinase inhibitor 1 | | Authors: | Wu, B, Arumugam, S, Semenchenko, V, Brew, K, Van Doren, S.R. | | Deposit date: | 1999-09-22 | | Release date: | 1999-12-22 | | Last modified: | 2022-03-23 | | Method: | SOLUTION NMR | | Cite: | NMR structure of tissue inhibitor of metalloproteinases-1 implicates localized induced fit in recognition of matrix metalloproteinases.
J.Mol.Biol., 295, 2000
|
|
5UE5
 
 | | proMMP-7 with heparin octasaccharide bound to the catalytic domain | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, CALCIUM ION, Matrilysin, ... | | Authors: | Fulcher, Y.G, Prior, S.H, Linhardt, R.J, Van Doren, S.R. | | Deposit date: | 2016-12-29 | | Release date: | 2017-07-19 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Glycan Activation of a Sheddase: Electrostatic Recognition between Heparin and proMMP-7.
Structure, 25, 2017
|
|
5UE2
 
 | | proMMP-7 with heparin octasaccharide bridging between domains | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, CALCIUM ION, Matrilysin, ... | | Authors: | Fulcher, Y.G, Prior, S.H, Linhardt, R.J, Van Doren, S.R. | | Deposit date: | 2016-12-29 | | Release date: | 2017-07-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Glycan Activation of a Sheddase: Electrostatic Recognition between Heparin and proMMP-7.
Structure, 25, 2017
|
|
7MY8
 
 | |
1FAF
 
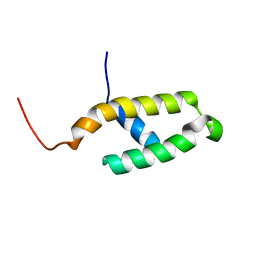 | | NMR STRUCTURE OF THE N-TERMINAL J DOMAIN OF MURINE POLYOMAVIRUS T ANTIGENS. | | Descriptor: | LARGE T ANTIGEN | | Authors: | Berjanskii, M.V, Riley, M.I, Xie, A, Semenchenko, V, Folk, W.R, Van Doren, S.R. | | Deposit date: | 2000-07-13 | | Release date: | 2000-11-22 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the N-terminal J domain of murine polyomavirus T antigens. Implications for DnaJ-like domains and for mutations of T antigens.
J.Biol.Chem., 275, 2000
|
|
2N8R
 
 | |
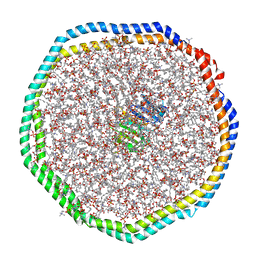
6CM1
 
 | |
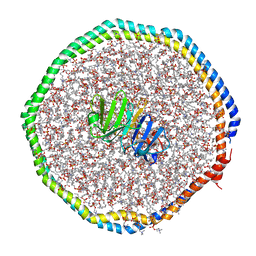
6CLZ
 
 | |
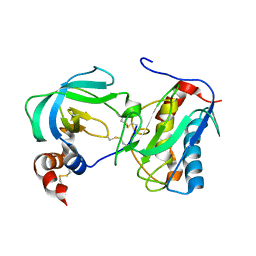
1OO9
 
 | |
1JP0
 
 | |
1JOX
 
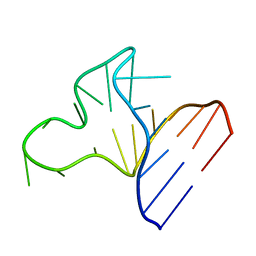 | |
1MZK
 
 | | NMR structure of kinase-interacting FHA domain of kinase associated protein phosphatase, KAPP in Arabidopsis | | Descriptor: | KINASE ASSOCIATED PROTEIN PHOSPHATASE | | Authors: | Lee, G, Ding, Z, Walker, J.C, Van Doren, S.R. | | Deposit date: | 2002-10-08 | | Release date: | 2003-09-09 | | Last modified: | 2022-12-21 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the forkhead-associated domain from the Arabidopsis receptor kinase-associated protein phosphatase.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
2MZH
 
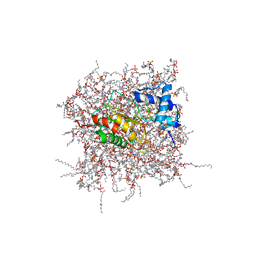 | | NMR Solution Structure of the PRO Form of Human Matrilysin (proMMP-7) in Complex with Zwitterionic Membrane | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, Matrilysin, ... | | Authors: | Prior, S.H, Van Doren, S.R. | | Deposit date: | 2015-02-12 | | Release date: | 2015-11-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Charge-Triggered Membrane Insertion of Matrix Metalloproteinase-7, Supporter of Innate Immunity and Tumors.
Structure, 23, 2015
|
|
2MZI
 
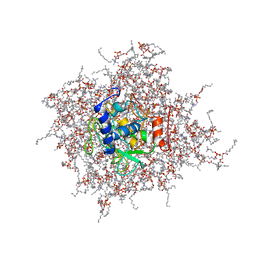 | | NMR Solution Structure of the PRO Form of Human Matrilysin (proMMP-7) in Complex with Anionic Membrane | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, CHOLEST-5-EN-3-YL HYDROGEN SULFATE, ... | | Authors: | Prior, S.H, Van Doren, S.R. | | Deposit date: | 2015-02-12 | | Release date: | 2016-09-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Charge-Triggered Membrane Insertion of Matrix Metalloproteinase-7, Supporter of Innate Immunity and Tumors.
Structure, 23, 2015
|
|
2MLS
 
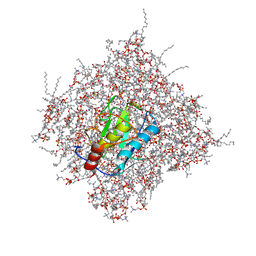 | | Membrane Bilayer complex with Matrix Metalloproteinase-12 at its Beta-face | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, Macrophage metalloelastase, ... | | Authors: | Koppisetti, R.K, Fulcher, Y.G, Prior, S.H, Lenoir, M, Overduin, M, Van Doren, S.R. | | Deposit date: | 2014-03-04 | | Release date: | 2014-12-03 | | Last modified: | 2014-12-10 | | Method: | SOLUTION NMR | | Cite: | Ambidextrous binding of cell and membrane bilayers by soluble matrix metalloproteinase-12.
Nat Commun, 5, 2014
|
|
2MLR
 
 | | Membrane Bilayer complex with Matrix Metalloproteinase-12 at its Alpha-face | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, Macrophage metalloelastase, ... | | Authors: | Koppisetti, R.K, Fulcher, Y.G, Prior, S.H, Lenoir, M, Overduin, M, Van Doren, S.R. | | Deposit date: | 2014-03-04 | | Release date: | 2014-12-03 | | Last modified: | 2014-12-10 | | Method: | SOLUTION NMR | | Cite: | Ambidextrous binding of cell and membrane bilayers by soluble matrix metalloproteinase-12.
Nat Commun, 5, 2014
|
|
2MQS
 
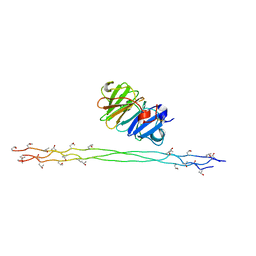 | |
2MZE
 
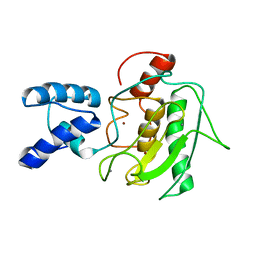 | | NMR Solution Structure of the PRO Form of Human Matrilysin (proMMP-7) | | Descriptor: | CALCIUM ION, Matrilysin, ZINC ION | | Authors: | Prior, S.H, Fulcher, Y.G, Van Doren, S.R. | | Deposit date: | 2015-02-11 | | Release date: | 2015-11-11 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Charge-Triggered Membrane Insertion of Matrix Metalloproteinase-7, Supporter of Innate Immunity and Tumors.
Structure, 23, 2015
|
|
2POJ
 
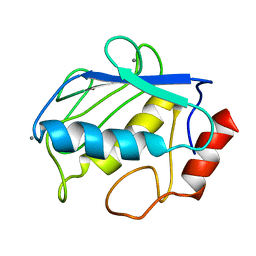 | |
3RSM
 
 | | Crystal structure of S108C mutant of PMM/PGM | | Descriptor: | PHOSPHATE ION, Phosphomannomutase/phosphoglucomutase, ZINC ION | | Authors: | Akella, A, Anbanandam, A, Kelm, A, Wei, Y, Mehra-Chaudhary, R, Beamer, L, Van Doren, S. | | Deposit date: | 2011-05-02 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Solution NMR of a 463-residue phosphohexomutase: domain 4 mobility, substates, and phosphoryl transfer defect.
Biochemistry, 51, 2012
|
|
1UEA
 
 | | MMP-3/TIMP-1 COMPLEX | | Descriptor: | CALCIUM ION, MATRIX METALLOPROTEINASE-3, TISSUE INHIBITOR OF METALLOPROTEINASE-1, ... | | Authors: | Bode, W, Maskos, K, Gomis-Rueth, F.-X, Nagase, H. | | Deposit date: | 1997-06-06 | | Release date: | 1998-10-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism of inhibition of the human matrix metalloproteinase stromelysin-1 by TIMP-1.
Nature, 389, 1997
|
|
