5J8E
 
 | |
1BG2
 
 | | HUMAN UBIQUITOUS KINESIN MOTOR DOMAIN | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, KINESIN, ... | | Authors: | Kull, F.J, Sablin, E.P, Lau, R, Fletterick, R.J, Vale, R.D. | | Deposit date: | 1998-06-04 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the kinesin motor domain reveals a structural similarity to myosin.
Nature, 380, 1996
|
|
4W8F
 
 | | Crystal structure of the dynein motor domain in the AMPPNP-bound state | | Descriptor: | Dynein heavy chain lysozyme chimera, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Cheng, H.-C, Bhabha, G, Zhang, N, Vale, R.D. | | Deposit date: | 2014-08-24 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.541 Å) | | Cite: | Allosteric communication in the Dynein motor domain.
Cell, 159, 2014
|
|
4W7G
 
 | |
3QMZ
 
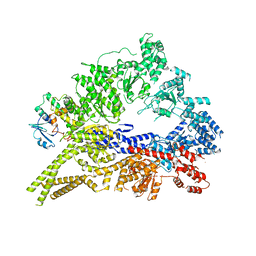 | | Crystal structure of the cytoplasmic dynein heavy chain motor domain | | Descriptor: | Cytoplasmic dynein heavy chain, Glutathione-S-transferase | | Authors: | Cho, C, Carter, A.P, Jin, L, Vale, R.D. | | Deposit date: | 2011-02-07 | | Release date: | 2011-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Crystal structure of the dynein motor domain.
Science, 331, 2011
|
|
8TO0
 
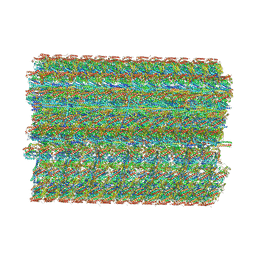 | | 48-nm repeating structure of doublets from mouse sperm flagella | | Descriptor: | Cilia- and flagella- associated protein 210, Cilia- and flagella-associated protein 107, Cilia- and flagella-associated protein 141, ... | | Authors: | Chen, Z, Shiozak, M, Hass, K.M, Skinner, W, Zhao, S, Guo, C, Polacco, B.J, Yu, Z, Krogan, N.J, Kaake, R.M, Vale, R.D, Agard, D.A. | | Deposit date: | 2023-08-02 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | De novo protein identification in mammalian sperm using in situ cryoelectron tomography and AlphaFold2 docking.
Cell, 186, 2023
|
|
7JR9
 
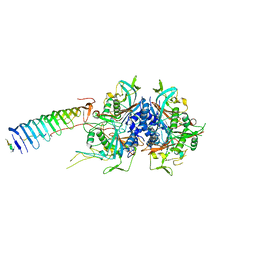 | | Chlamydomonas reinhardtii radial spoke minimal head complex | | Descriptor: | Flagellar radial spoke protein 10, Flagellar radial spoke protein 4, Flagellar radial spoke protein 6, ... | | Authors: | Grossman-Haham, I, Coudray, N, Yu, Z, Wang, F, Bhabha, G, Vale, R.D. | | Deposit date: | 2020-08-11 | | Release date: | 2020-12-16 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structure of the radial spoke head and insights into its role in mechanoregulation of ciliary beating.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7JRJ
 
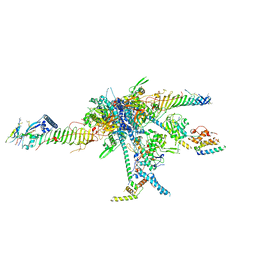 | | Chlamydomonas reinhardtii radial spoke head and neck (recombinant) | | Descriptor: | Flagellar radial spoke protein 1, Flagellar radial spoke protein 10, Flagellar radial spoke protein 2, ... | | Authors: | Grossman-Haham, I, Coudray, N, Yu, Z, Wang, F, Zhang, N, Bhabha, G, Vale, R.D. | | Deposit date: | 2020-08-12 | | Release date: | 2020-12-16 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structure of the radial spoke head and insights into its role in mechanoregulation of ciliary beating.
Nat.Struct.Mol.Biol., 28, 2021
|
|
1YIB
 
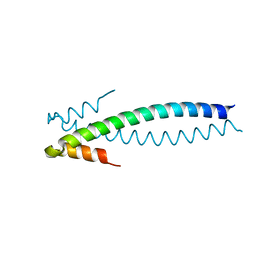 | | Crystal Structure of the Human EB1 C-terminal Dimerization Domain | | Descriptor: | Microtubule-associated protein RP/EB family member 1 | | Authors: | Slep, K.C, Rogers, S.L, Elliott, S.L, Ohkura, H, Kolodziej, P.A, Vale, R.D. | | Deposit date: | 2005-01-11 | | Release date: | 2005-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural determinants for EB1-mediated recruitment of APC and spectraplakins to the microtubule plus end
J.Cell Biol., 168, 2005
|
|
1YIG
 
 | | Crystal Structure of the Human EB1 C-terminal Dimerization Domain | | Descriptor: | Microtubule-associated protein RP/EB family member 1 | | Authors: | Slep, K.C, Rogers, S.L, Elliott, S.L, Ohkura, H, Kolodziej, P.A, Vale, R.D. | | Deposit date: | 2005-01-11 | | Release date: | 2005-03-08 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural determinants for EB1-mediated recruitment of APC and spectraplakins to the microtubule plus end
J.Cell Biol., 168, 2005
|
|
3B9P
 
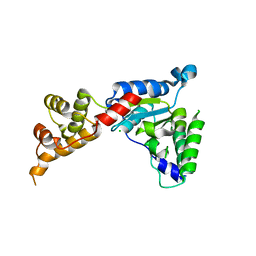 | | Spastin | | Descriptor: | CG5977-PA, isoform A, CHLORIDE ION | | Authors: | Roll-Mecak, A, Vale, R.D. | | Deposit date: | 2007-11-06 | | Release date: | 2008-01-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of microtubule severing by the hereditary spastic paraplegia protein spastin.
Nature, 451, 2008
|
|
2NCD
 
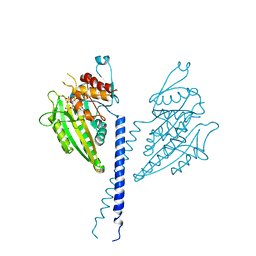 | | NCD (NON-CLARET DISJUNCTIONAL) DIMER FROM D. MELANOGASTER | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PROTEIN (Kinesin motor NCD), SULFATE ION | | Authors: | Sablin, E.P, Case, R.B, Dai, S.C, Hart, C.L, Ruby, A, Vale, R.D, Fletterick, R.J. | | Deposit date: | 1999-06-04 | | Release date: | 1999-06-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Direction determination in the minus-end-directed kinesin motor ncd.
Nature, 395, 1998
|
|
2QK0
 
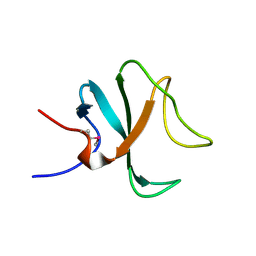 | |
2QK2
 
 | |
2QJX
 
 | |
2QJZ
 
 | |
2QK1
 
 | |
3ERR
 
 | |
6BJ3
 
 | | TCR55 in complex with HIV(Pol448-456)/HLA-B35 | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, HIV Pol B35 peptide, ... | | Authors: | Jude, K.M, Sibener, L.V, Garcia, K.C. | | Deposit date: | 2017-11-03 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Isolation of a Structural Mechanism for Uncoupling T Cell Receptor Signaling from Peptide-MHC Binding.
Cell, 174, 2018
|
|
6BJ8
 
 | | TCR55 in complex with Pep20/HLA-B35 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Jude, K.M, Sibener, L.V, Yang, X, Garcia, K.C. | | Deposit date: | 2017-11-05 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Isolation of a Structural Mechanism for Uncoupling T Cell Receptor Signaling from Peptide-MHC Binding.
Cell, 174, 2018
|
|
6BJ2
 
 | | TCR589 in complex with HIV(Pol448-456)/HLA-B35 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, ... | | Authors: | Jude, K.M, Sibener, L.V, Garcia, K.C. | | Deposit date: | 2017-11-03 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Isolation of a Structural Mechanism for Uncoupling T Cell Receptor Signaling from Peptide-MHC Binding.
Cell, 174, 2018
|
|
5UWA
 
 | | Structure of E. coli phospholipid binding protein MlaC | | Descriptor: | (2S)-3-(2-aminoethoxy)propane-1,2-diyl dihexadecanoate, Probable phospholipid-binding protein MlaC | | Authors: | Bhabha, G, Ekiert, D.C. | | Deposit date: | 2017-02-20 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Architectures of Lipid Transport Systems for the Bacterial Outer Membrane.
Cell, 169, 2017
|
|
5UWB
 
 | |
5UVN
 
 | |
5UW2
 
 | |
