4WKM
 
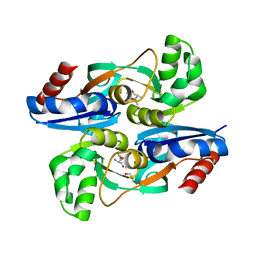 | | AmpR effector binding domain from Citrobacter freundii bound to UDP-MurNAc-pentapeptide | | Descriptor: | ALA-FGA-API-DAL-DAL, GLYCEROL, LysR family transcriptional regulator, ... | | Authors: | Vadlamani, G, Reeve, T.M, Mark, B.L. | | Deposit date: | 2014-10-02 | | Release date: | 2014-12-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The beta-Lactamase Gene Regulator AmpR Is a Tetramer That Recognizes and Binds the d-Ala-d-Ala Motif of Its Repressor UDP-N-acetylmuramic Acid (MurNAc)-pentapeptide.
J.Biol.Chem., 290, 2015
|
|
7MPY
 
 | |
5UTQ
 
 | |
5UTP
 
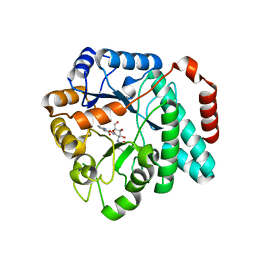 | | Crystal structure of Burkholderia cenocepacia family 3 glycoside hydrolase (NagZ) bound to N-ethylbutyryl-PUGNAc | | Descriptor: | Beta-hexosaminidase, N-[(2Z,3R,4R,5S,6R)-4,5-dihydroxy-6-(hydroxymethyl)-2-{[(phenylcarbamoyl)oxy]imino}tetrahydro-2H-pyran-3-yl]-2-ethylbutanamide | | Authors: | Vadlamani, G, Mark, B.L. | | Deposit date: | 2017-02-15 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational flexibility of the glycosidase NagZ allows it to bind structurally diverse inhibitors to suppress beta-lactam antibiotic resistance.
Protein Sci., 26, 2017
|
|
5UTR
 
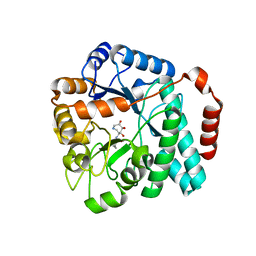 | | Crystal structure of Burkholderia cenocepacia family 3 glycoside hydrolase (NagZ) bound to (3S,4R,5R,6S)-3-butyryl-4,5,6-trihydroxyazepane | | Descriptor: | Beta-hexosaminidase, N-[(3S,4R,5R,6S)-4,5,6-trihydroxyazepan-3-yl]butanamide | | Authors: | Vadlamani, G, Mark, B.L. | | Deposit date: | 2017-02-15 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Conformational flexibility of the glycosidase NagZ allows it to bind structurally diverse inhibitors to suppress beta-lactam antibiotic resistance.
Protein Sci., 26, 2017
|
|
4MSS
 
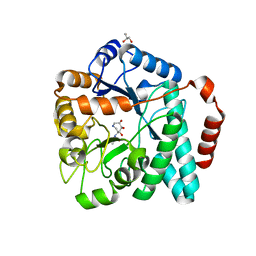 | | Crystal structure of Burkholderia cenocepacia family 3 glycoside hydrolase (NagZ) bound to (3S,4R,5R,6S)-3-acetamido-4,5,6-trihydroxyazepane | | Descriptor: | Beta-hexosaminidase 1, GLYCEROL, N-[(3S,4R,5R,6S)-4,5,6-trihydroxyazepan-3-yl]acetamide | | Authors: | Vadlamani, G, Mark, B.L. | | Deposit date: | 2013-09-18 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Selective trihydroxyazepane NagZ inhibitors increase sensitivity of Pseudomonas aeruginosa to beta-lactams.
Chem.Commun.(Camb.), 49, 2013
|
|
5FD0
 
 | |
5FCZ
 
 | |
7MMY
 
 | |
6MKQ
 
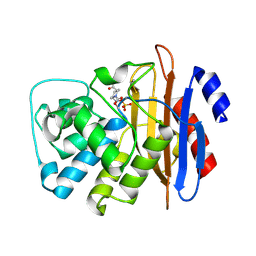 | | Carbapenemase VCC-1 bound to avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase | | Authors: | Mark, B.L, Vadlamani, G. | | Deposit date: | 2018-09-26 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Basis for the Potent Inhibition of the Emerging Carbapenemase VCC-1 by Avibactam.
Antimicrob. Agents Chemother., 63, 2019
|
|
6MK6
 
 | |
6WQL
 
 | |
6WQJ
 
 | |
7L51
 
 | |
7L54
 
 | |
7L53
 
 | |
7L55
 
 | |
7M3U
 
 | |
5VTG
 
 | |
