4QCD
 
 | | Neutron crystal structure of phycocyanobilin:ferredoxin oxidoreductase in complex with biliverdin IXalpha at room temperature. | | Descriptor: | BILIVERDINE IX ALPHA, Phycocyanobilin:ferredoxin oxidoreductase, trideuteriooxidanium | | Authors: | Unno, M, Ishikawa-Suto, K, Ishihara, M, Hagiwara, Y, Sugishima, M, Wada, K, Fukuyama, K. | | Deposit date: | 2014-05-10 | | Release date: | 2015-04-29 | | Last modified: | 2024-03-20 | | Method: | NEUTRON DIFFRACTION (1.932 Å), X-RAY DIFFRACTION | | Cite: | Insights into the Proton Transfer Mechanism of a Bilin Reductase PcyA Following Neutron Crystallography.
J. Am. Chem. Soc., 137, 2015
|
|
3QXM
 
 | | Crystal Structure of Human GluK2 Ligand-Binding Core in Complex with Novel Marine-Derived Toxins, Neodysiherbaine A | | Descriptor: | (2R,3aR,6R,7R,7aR)-2-[(2S)-2-amino-2-carboxyethyl]-6,7-dihydroxyhexahydro-2H-furo[3,2-b]pyran-2-carboxylic acid, Glutamate receptor ionotropic, kainate 2 | | Authors: | Unno, M, Sasaki, M, Ikeda-Saito, M. | | Deposit date: | 2011-03-02 | | Release date: | 2011-10-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Binding and Selectivity of the Marine Toxin Neodysiherbaine A and Its Synthetic Analogues to GluK1 and GluK2 Kainate Receptors.
J.Mol.Biol., 413, 2011
|
|
3FUZ
 
 | | Crystal structure of the human glutamate receptor, GluR5, ligand-binding core in complex with L-glutamate in space group P1 | | Descriptor: | GLUTAMIC ACID, Glutamate receptor, ionotropic kainate 1, ... | | Authors: | Unno, M, Sasaki, M, Ikeda-Saito, M. | | Deposit date: | 2009-01-15 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Binding and Selectivity of the Marine Toxin Neodysiherbaine A and Its Synthetic Analogues to GluK1 and GluK2 Kainate Receptors.
J.Mol.Biol., 413, 2011
|
|
3FVK
 
 | | Crystal structure of the human glutamate receptor, GluR5, ligand-binding core in complex with 8-deoxy-neodysiherbaine A in space group P1 | | Descriptor: | (2R,3aR,6S,7aR)-2-[(2S)-2-amino-3-hydroxy-3-oxo-propyl]-6-hydroxy-3,3a,5,6,7,7a-hexahydrofuro[4,5-b]pyran-2-carboxylic acid, GLYCEROL, Glutamate receptor, ... | | Authors: | Unno, M, Sasaki, M, Ikeda-Saito, M. | | Deposit date: | 2009-01-15 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding and Selectivity of the Marine Toxin Neodysiherbaine A and Its Synthetic Analogues to GluK1 and GluK2 Kainate Receptors.
J.Mol.Biol., 413, 2011
|
|
3FVO
 
 | | Crystal structure of the human glutamate receptor, GluR5, ligand-binding core in complex with 8-epi-neodysiherbaine A in space group P1 | | Descriptor: | (2R,3aR,6R,7S,7aR)-2-[(2S)-2-amino-3-hydroxy-3-oxo-propyl]-6,7-dihydroxy-3,3a,5,6,7,7a-hexahydrofuro[4,5-b]pyran-2-carboxylic acid, GLYCEROL, Glutamate receptor, ... | | Authors: | Unno, M, Sasaki, M, Ikeda-Saito, M. | | Deposit date: | 2009-01-16 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structures of Human GluR5 Ligand-Binding Core in Complexes with Novel Marine-Derived Toxins, Dysiherbaine and Neodysiherbaine A, and Their Analogues
To be Published
|
|
3FVN
 
 | | Crystal structure of the human glutamate receptor, GluR5, ligand-binding core in complex with 9-deoxy-neodysiherbaine A in space group P1 | | Descriptor: | (2R,3aR,7R,7aR)-2-[(2S)-2-amino-3-hydroxy-3-oxo-propyl]-7-hydroxy-3,3a,5,6,7,7a-hexahydrofuro[4,5-b]pyran-2-carboxylic acid, GLYCEROL, Glutamate receptor, ... | | Authors: | Unno, M, Sasaki, M, Ikeda-Saito, M. | | Deposit date: | 2009-01-16 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding and Selectivity of the Marine Toxin Neodysiherbaine A and Its Synthetic Analogues to GluK1 and GluK2 Kainate Receptors.
J.Mol.Biol., 413, 2011
|
|
3FV2
 
 | | Crystal structure of the human glutamate receptor, GluR5, ligand-binding core in complex with neodysiherbaine A in space group P1 | | Descriptor: | (2R,3aR,6R,7R,7aR)-2-[(2S)-2-amino-2-carboxyethyl]-6,7-dihydroxyhexahydro-2H-furo[3,2-b]pyran-2-carboxylic acid, GLYCEROL, Glutamate receptor, ... | | Authors: | Unno, M, Sasaki, M, Ikeda-Saito, M. | | Deposit date: | 2009-01-15 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding and Selectivity of the Marine Toxin Neodysiherbaine A and Its Synthetic Analogues to GluK1 and GluK2 Kainate Receptors.
J.Mol.Biol., 413, 2011
|
|
3FV1
 
 | | Crystal Structure of the human glutamate receptor, GluR5, ligand-binding core in complex with dysiherbaine in space group P1 | | Descriptor: | (2R,3aR,6S,7R,7aR)-2-[(2S)-2-amino-2-carboxyethyl]-6-hydroxy-7-(methylamino)hexahydro-2H-furo[3,2-b]pyran-2-carboxylic acid, GLYCEROL, Glutamate receptor, ... | | Authors: | Unno, M, Sasaki, M, Ikeda-Saito, M. | | Deposit date: | 2009-01-15 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding and Selectivity of the Marine Toxin Neodysiherbaine A and Its Synthetic Analogues to GluK1 and GluK2 Kainate Receptors.
J.Mol.Biol., 413, 2011
|
|
3FVG
 
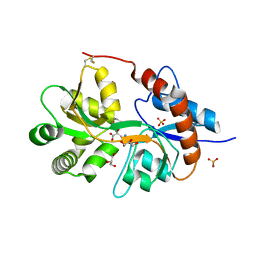 | | Crystal structure of the human glutamate receptor, GluR5, ligand-binding core in complex with MSVIII-19 in space group P1 | | Descriptor: | (2R,3aR,7aR)-2-[(2S)-2-amino-3-hydroxy-3-oxo-propyl]-3,3a,5,6,7,7a-hexahydrofuro[4,5-b]pyran-2-carboxylic acid, GLYCEROL, Glutamate receptor, ... | | Authors: | Unno, M, Sasaki, M, Ikeda-Saito, M. | | Deposit date: | 2009-01-15 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding and Selectivity of the Marine Toxin Neodysiherbaine A and Its Synthetic Analogues to GluK1 and GluK2 Kainate Receptors.
J.Mol.Biol., 413, 2011
|
|
4GPH
 
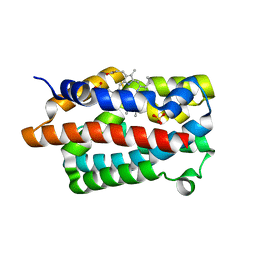 | |
4GPC
 
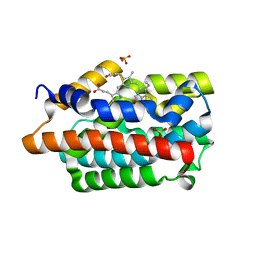 | |
4GOH
 
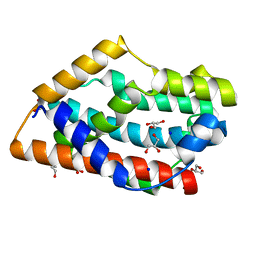 | |
4GPF
 
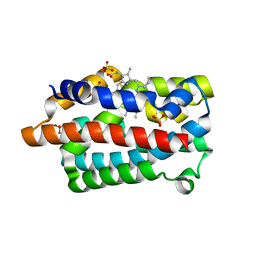 | |
1V8X
 
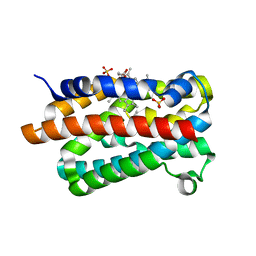 | | Crystal Structure of the Dioxygen-bound Heme Oxygenase from Corynebacterium diphtheriae | | Descriptor: | Heme oxygenase, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Unno, M, Matsui, T, Chu, G.C, Couture, M, Yoshida, T, Rousseau, D.L, Olson, J.S, Ikeda-Saito, M. | | Deposit date: | 2004-01-15 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Dioxygen-bound Heme Oxygenase from Corynebacterium diphtheriae: IMPLICATIONS FOR HEME OXYGENASE FUNCTION.
J.Biol.Chem., 279, 2004
|
|
1IRU
 
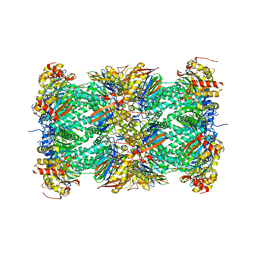 | | Crystal Structure of the mammalian 20S proteasome at 2.75 A resolution | | Descriptor: | 20S proteasome, MAGNESIUM ION | | Authors: | Unno, M, Mizushima, T, Morimoto, Y, Tomisugi, Y, Tanaka, K, Yasuoka, N, Tsukihara, T. | | Deposit date: | 2001-10-24 | | Release date: | 2002-05-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The structure of the mammalian 20S proteasome at 2.75 A resolution.
Structure, 10, 2002
|
|
3NSI
 
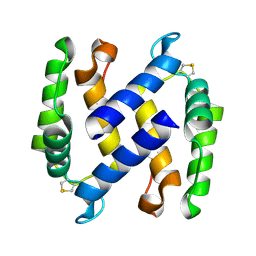 | |
3NSK
 
 | |
3NSL
 
 | |
3NSO
 
 | |
2ZNS
 
 | | Crystal structure of the ligand-binding core of the human ionotropic glutamate receptor, GluR5, in complex with glutamate | | Descriptor: | GLUTAMIC ACID, Glutamate receptor, ionotropic kainate 1 | | Authors: | Unno, M, Sasaki, M, Ikeda-Saito, M. | | Deposit date: | 2008-05-01 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding and Selectivity of the Marine Toxin Neodysiherbaine A and Its Synthetic Analogues to GluK1 and GluK2 Kainate Receptors.
J.Mol.Biol., 413, 2011
|
|
2ZNU
 
 | | Crystal structure of the ligand-binding core of the human ionotropic glutamate receptor, GluR5, in complex with a novel selective agonist, neodysiherbaine A | | Descriptor: | (2R,3aR,6R,7R,7aR)-2-[(2S)-2-amino-2-carboxyethyl]-6,7-dihydroxyhexahydro-2H-furo[3,2-b]pyran-2-carboxylic acid, BETA-MERCAPTOETHANOL, Glutamate receptor, ... | | Authors: | Unno, M, Sasaki, M, Ikeda-Saito, M. | | Deposit date: | 2008-05-01 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding and Selectivity of the Marine Toxin Neodysiherbaine A and Its Synthetic Analogues to GluK1 and GluK2 Kainate Receptors.
J.Mol.Biol., 413, 2011
|
|
2ZNT
 
 | | Crystal structure of the ligand-binding core of the human ionotropic glutamate receptor, GluR5, in complex with a novel selective agonist, dysiherbaine | | Descriptor: | (2R,3aR,6S,7R,7aR)-2-[(2S)-2-amino-2-carboxyethyl]-6-hydroxy-7-(methylamino)hexahydro-2H-furo[3,2-b]pyran-2-carboxylic acid, BETA-MERCAPTOETHANOL, Glutamate receptor, ... | | Authors: | Unno, M, Sasaki, M, Ikeda-Saito, M. | | Deposit date: | 2008-05-01 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Binding and Selectivity of the Marine Toxin Neodysiherbaine A and Its Synthetic Analogues to GluK1 and GluK2 Kainate Receptors.
J.Mol.Biol., 413, 2011
|
|
2Z6T
 
 | | Crystal structure of the ferric peroxo myoglobin | | Descriptor: | Myoglobin, PEROXIDE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Unno, M, Kusama, S, Chen, H, Shaik, S, Ikeda-Saito, M. | | Deposit date: | 2007-08-08 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Characterization of the Fleeting Ferric Peroxo Species in Myoglobin: Experiment and Theory
J.Am.Chem.Soc., 129, 2007
|
|
2Z6S
 
 | | Crystal structure of the oxy myoglobin free from X-ray-induced photoreduction | | Descriptor: | Myoglobin, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Unno, M, Kusama, S, Chen, H, Shaik, S, Ikeda-Saito, M. | | Deposit date: | 2007-08-08 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Characterization of the Fleeting Ferric Peroxo Species in Myoglobin: Experiment and Theory
J.Am.Chem.Soc., 129, 2007
|
|
7YL8
 
 | |
