1KBD
 
 | | SOLUTION STRUCTURE OF A 16 BASE-PAIR DNA RELATED TO THE HIV-1 KAPPA B SITE | | Descriptor: | DNA (5'-D(*CP*CP*TP*GP*GP*AP*AP*AP*GP*TP*CP*CP*CP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*GP*GP*GP*AP*CP*TP*TP*TP*CP*CP*AP*GP*G)-3') | | Authors: | Tisne, C, Hantz, E, Hartmann, B, Delepierre, M. | | Deposit date: | 1998-11-28 | | Release date: | 1998-12-02 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a non-palindromic 16 base-pair DNA related to the HIV-1 kappa B site: evidence for BI-BII equilibrium inducing a global dynamic curvature of the duplex.
J.Mol.Biol., 279, 1998
|
|
3KBD
 
 | | MUTATED NF KAPPA-B SITE, BI MODEL | | Descriptor: | DNA (5'-D(*CP*CP*TP*GP*GP*AP*AP*AP*GP*TP*GP*AP*GP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*CP*TP*CP*AP*CP*TP*TP*TP*CP*CP*AP*GP*G)-3') | | Authors: | Tisne, C, Hartmann, B, Delepierre, M. | | Deposit date: | 1998-11-30 | | Release date: | 1999-10-14 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NF-kappa B binding mechanism: a nuclear magnetic resonance and modeling study of a GGG --> CTC mutation.
Biochemistry, 38, 1999
|
|
2KBD
 
 | | 5'-D(*CP*TP*GP*GP*GP*GP*AP*CP*TP*TP*TP*CP*CP*AP*GP*G)-3', 5'-D(*CP*CP*TP*GP*GP*AP*AP*AP*GP*TP*CP*CP*CP*CP*AP*G)-3' | | Descriptor: | DNA (5'-D(*CP*CP*TP*GP*GP*AP*AP*AP*GP*TP*CP*CP*CP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*GP*GP*GP*AP*CP*TP*TP*TP*CP*CP*AP*GP*G)-3') | | Authors: | Tisne, C, Hantz, E, Hartmann, B, Delepierre, M. | | Deposit date: | 1998-12-07 | | Release date: | 1998-12-09 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a non-palindromic 16 base-pair DNA related to the HIV-1 kappa B site: evidence for BI-BII equilibrium inducing a global dynamic curvature of the duplex.
J.Mol.Biol., 279, 1998
|
|
4KBD
 
 | | DNA STRUCTURE OF A MUTATED KB SITE | | Descriptor: | DNA (5'-D(*CP*CP*TP*GP*GP*AP*AP*AP*GP*TP*GP*AP*GP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*CP*TP*CP*AP*CP*TP*TP*TP*CP*CP*AP*GP*G)-3') | | Authors: | Tisne, C, Hartmann, B, Delepierre, M. | | Deposit date: | 1998-11-30 | | Release date: | 1999-10-14 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NF-kappa B binding mechanism: a nuclear magnetic resonance and modeling study of a GGG --> CTC mutation.
Biochemistry, 38, 1999
|
|
6QE6
 
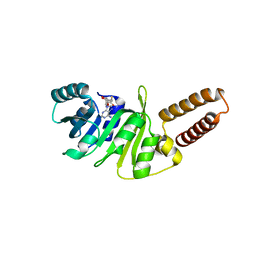 | | Structure of M. capricolum TrmK in complex with the natural cofactor product S-adenosyl-homocysteine (SAH) | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, tRNA (Adenine(22)-N(1))-methyltransferase | | Authors: | Oerum, S, Catala, M, Atdjian, C, Brachet, F, Ponchon, L, Barraud, P, Iannazzo, L, Droogmans, L, Braud, E, Etheve-Quelquejeu, M, Tisne, C. | | Deposit date: | 2019-01-04 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Bisubstrate analogues as structural tools to investigate m6A methyltransferase active sites.
Rna Biol., 16, 2019
|
|
6QE5
 
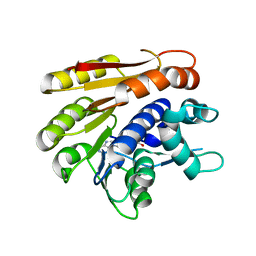 | | Structure of E.coli RlmJ in complex with the natural cofactor product S-adenosyl-homocysteine (SAH) | | Descriptor: | Ribosomal RNA large subunit methyltransferase J, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Oerum, S, Catala, M, Atdjian, C, Brachet, F, Ponchon, L, Barraud, P, Iannazzo, L, Droogmans, L, Braud, E, Etheve-Quelquejeu, M, Tisne, C. | | Deposit date: | 2019-01-04 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Bisubstrate analogues as structural tools to investigate m6A methyltransferase active sites.
Rna Biol., 16, 2019
|
|
2PWY
 
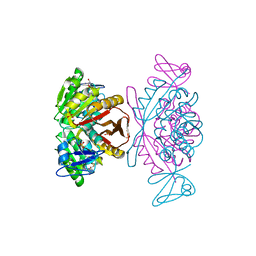 | | Crystal Structure of a m1A58 tRNA methyltransferase | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, tRNA (adenine-N(1)-)-methyltransferase | | Authors: | Barraud, P, Golinelli-Pimpaneau, B, Tisne, C. | | Deposit date: | 2007-05-14 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Thermus thermophilus tRNA m(1)A(58) Methyltransferase and Biophysical Characterization of Its Interaction with tRNA.
J.Mol.Biol., 377, 2008
|
|
7P8Q
 
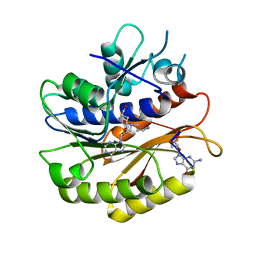 | | Structure of E.coli RlmJ in complex with an RNA conjugate (GA-SAM) | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl](3-aminopropyl)amino}-5'-deoxyadenosine, RNA conjugate (GA-SAM), Ribosomal RNA large subunit methyltransferase J | | Authors: | Meynier, V, Catala, M, Oerum, S, Barraud, P, Tisne, C. | | Deposit date: | 2021-07-23 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.289 Å) | | Cite: | Synthesis of RNA-cofactor conjugates and structural exploration of RNA recognition by an m6A RNA methyltransferase.
Nucleic Acids Res., 50, 2022
|
|
7P9I
 
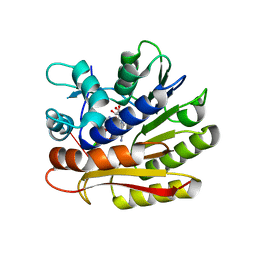 | | Structure of E.coli RlmJ in complex with an RNA conjugate (GAA-SAM) | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl](3-aminopropyl)amino}-5'-deoxyadenosine, RNA conjugate (GAA-SAM), Ribosomal RNA large subunit methyltransferase J | | Authors: | Meynier, V, Catala, M, Oerum, S, Barraud, P, Tisne, C. | | Deposit date: | 2021-07-27 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.594 Å) | | Cite: | Synthesis of RNA-cofactor conjugates and structural exploration of RNA recognition by an m6A RNA methyltransferase.
Nucleic Acids Res., 50, 2022
|
|
6YPB
 
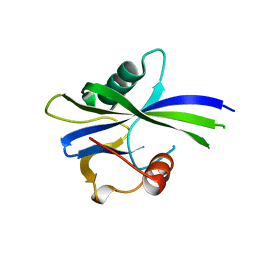 | | NUDIX1 hydrolase from Rosa x hybrida | | Descriptor: | Geranyl diphosphate phosphohydrolase | | Authors: | Degut, C, Rety, S, Tisne, C, Baudino, S. | | Deposit date: | 2020-04-15 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Functional diversification in the Nudix hydrolase gene family drives sesquiterpene biosynthesis in Rosa × wichurana.
Plant J., 104, 2020
|
|
6YPF
 
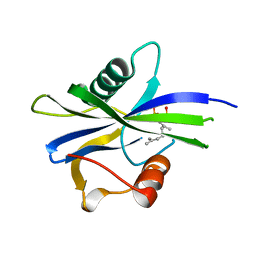 | | NUDIX1 hydrolase from Rosa x hybrida in complex with geranyl pyrophosphate | | Descriptor: | GERANYL DIPHOSPHATE, Geranyl diphosphate phosphohydrolase | | Authors: | Degut, C, Rety, S, Tisne, C, Baudino, S. | | Deposit date: | 2020-04-16 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Functional diversification in the Nudix hydrolase gene family drives sesquiterpene biosynthesis in Rosa × wichurana.
Plant J., 104, 2020
|
|
5C1I
 
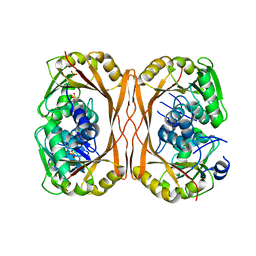 | | m1A58 tRNA methyltransferase mutant - D170A | | Descriptor: | SULFATE ION, tRNA (adenine(58)-N(1))-methyltransferase TrmI | | Authors: | Ponchon, L, Degut, C, Folly-Klan, M, Barraud, P, Tisne, C. | | Deposit date: | 2015-06-14 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The m1A58 modification in eubacterial tRNA: An overview of tRNA recognition and mechanism of catalysis by TrmI.
Biophys.Chem., 210, 2016
|
|
5C0O
 
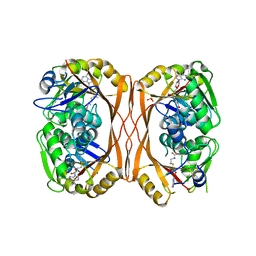 | | m1A58 tRNA methyltransferase mutant - Y78A | | Descriptor: | S-ADENOSYLMETHIONINE, SULFATE ION, tRNA (adenine(58)-N(1))-methyltransferase TrmI | | Authors: | Degut, C, Ponchon, L, Folly-Klan, M, Barraud, P, Tisne, C. | | Deposit date: | 2015-06-12 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | The m1A58 modification in eubacterial tRNA: An overview of tRNA recognition and mechanism of catalysis by TrmI.
Biophys.Chem., 210, 2016
|
|
6Z2B
 
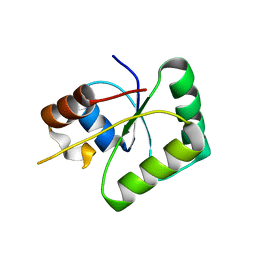 | |
7P9O
 
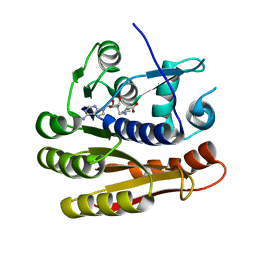 | | Structure of E.coli RlmJ in complex with a SAM analogue (CA) | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl]amino}-5'-deoxyadenosine, Ribosomal RNA large subunit methyltransferase J | | Authors: | Meynier, V, Catala, M, Oerum, S, Barraud, P, Tisne, C. | | Deposit date: | 2021-07-27 | | Release date: | 2022-05-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | Synthesis of RNA-cofactor conjugates and structural exploration of RNA recognition by an m6A RNA methyltransferase.
Nucleic Acids Res., 50, 2022
|
|
6Q56
 
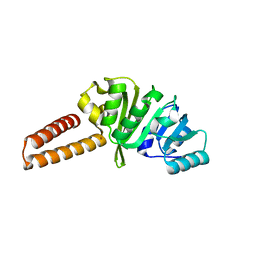 | | Crystal structure of the B. subtilis M1A22 tRNA methyltransferase TrmK | | Descriptor: | NICKEL (II) ION, tRNA (adenine(22)-N(1))-methyltransferase | | Authors: | Degut, C, Roovers, M, Barraud, P, Brachet, F, Feller, A, Larue, V, Al Refaii, A, Caillet, J, Droogmans, L, Tisne, C. | | Deposit date: | 2018-12-07 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of B. subtilis m1A22 tRNA methyltransferase TrmK: insights into tRNA recognition.
Nucleic Acids Res., 47, 2019
|
|
3CW6
 
 | | E. coli Initiator tRNA | | Descriptor: | Initiator tRNA | | Authors: | Barraud, P, Schmitt, E, Mechulam, Y, Dardel, F, Tisne, C. | | Deposit date: | 2008-04-21 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A unique conformation of the anticodon stem-loop is associated with the capacity of tRNAfMet to initiate protein synthesis.
Nucleic Acids Res., 36, 2008
|
|
3CW5
 
 | | E. coli Initiator tRNA | | Descriptor: | Initiator tRNA | | Authors: | Barraud, P, Schmitt, E, Mechulam, Y, Dardel, F, Tisne, C. | | Deposit date: | 2008-04-21 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A unique conformation of the anticodon stem-loop is associated with the capacity of tRNAfMet to initiate protein synthesis.
Nucleic Acids Res., 36, 2008
|
|
4JZT
 
 | | Crystal structure of the Bacillus subtilis pyrophosphohydrolase BsRppH (E68A mutant) bound to GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, dGTP pyrophosphohydrolase | | Authors: | Piton, J, Larue, V, Thillier, Y, Dorleans, A, Pellegrini, O, Li de la Sierra-Gallay, I, Vasseur, J.J, Debart, F, Tisne, C, Condon, C. | | Deposit date: | 2013-04-03 | | Release date: | 2013-05-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Bacillus subtilis RNA deprotection enzyme RppH recognizes guanosine in the second position of its substrates.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JZV
 
 | | Crystal structure of the Bacillus subtilis pyrophosphohydrolase BsRppH bound to a non-hydrolysable triphosphorylated dinucleotide RNA (pcp-pGpG) - second guanosine residue in guanosine binding pocket | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, RNA (5'-R(*(GCP)P*G)-3'), ... | | Authors: | Piton, J, Larue, V, Thillier, Y, Dorleans, A, Pellegrini, O, Li de la Sierra-Gallay, I, Vasseur, J.J, Debart, F, Tisne, C, Condon, C. | | Deposit date: | 2013-04-03 | | Release date: | 2013-05-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bacillus subtilis RNA deprotection enzyme RppH recognizes guanosine in the second position of its substrates.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JZS
 
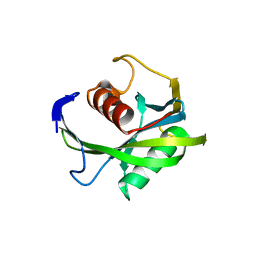 | | Crystal structure of the Bacillus subtilis pyrophosphohydrolase BsRppH (E68A mutant) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, dGTP pyrophosphohydrolase | | Authors: | Piton, J, Larue, V, Thillier, Y, Dorleans, A, Pellegrini, O, Li de la Sierra-Gallay, I, Vasseur, J.J, Debart, F, Tisne, C, Condon, C. | | Deposit date: | 2013-04-03 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bacillus subtilis RNA deprotection enzyme RppH recognizes guanosine in the second position of its substrates.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JZU
 
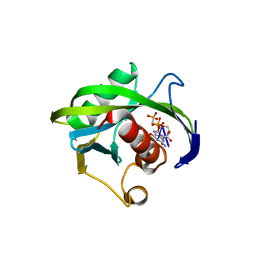 | | Crystal structure of the Bacillus subtilis pyrophosphohydrolase BsRppH bound to a non-hydrolysable triphosphorylated dinucleotide RNA (pcp-pGpG) - first guanosine residue in guanosine binding pocket | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, RNA (5'-R(*(GCP)P*G)-3'), RNA PYROPHOSPHOHYDROLASE | | Authors: | Piton, J, Larue, V, Thillier, Y, Dorleans, A, Pellegrini, O, Li de la Sierra-Gallay, I, Vasseur, J.J, Debart, F, Tisne, C, Condon, C. | | Deposit date: | 2013-04-03 | | Release date: | 2013-05-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Bacillus subtilis RNA deprotection enzyme RppH recognizes guanosine in the second position of its substrates.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6TNN
 
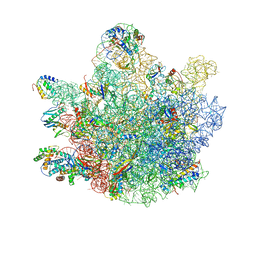 | | Mini-RNase III (Mini-III) bound to 50S ribosome with precursor 23S rRNA | | Descriptor: | 50S ribosomal protein L10, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Oerum, S, Dendooven, T, Gilet, L, Catala, M, Degut, C, Trinquier, A, Barraud, P, Luisi, B, Condon, C, Tisne, C. | | Deposit date: | 2019-12-09 | | Release date: | 2020-09-30 | | Last modified: | 2020-10-28 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structures of B. subtilis Maturation RNases Captured on 50S Ribosome with Pre-rRNAs.
Mol.Cell, 80, 2020
|
|
6TPQ
 
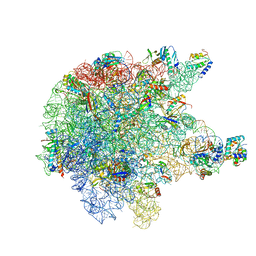 | | RNase M5 bound to 50S ribosome with precursor 5S rRNA | | Descriptor: | 50S ribosomal protein L10, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Oerum, S, Dendooven, T, Gilet, L, Catala, M, Degut, C, Trinquier, A, Barraud, P, Luisi, B, Condon, C, Tisne, C. | | Deposit date: | 2019-12-13 | | Release date: | 2020-09-30 | | Last modified: | 2020-10-28 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structures of B. subtilis Maturation RNases Captured on 50S Ribosome with Pre-rRNAs.
Mol.Cell, 80, 2020
|
|
6TGJ
 
 | |
