1Z3I
 
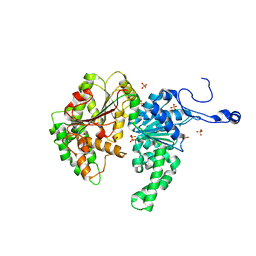 | | Structure of the SWI2/SNF2 chromatin remodeling domain of eukaryotic Rad54 | | Descriptor: | SULFATE ION, ZINC ION, similar to RAD54-like | | Authors: | Thoma, N.H, Czyzewski, B.K, Alexeev, A.A, Mazin, A.V, Kowalczykowski, S.C, Pavletich, N.P. | | Deposit date: | 2005-03-12 | | Release date: | 2005-04-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the SWI2/SNF2 chromatin-remodeling domain of eukaryotic Rad54.
Nat.Struct.Mol.Biol., 12, 2005
|
|
5REQ
 
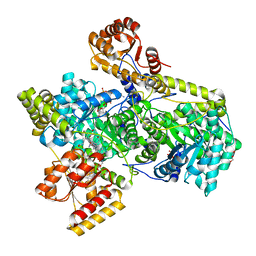 | | Methylmalonyl-COA MUTASE, Y89F Mutant, substrate complex | | Descriptor: | COBALAMIN, GLYCEROL, METHYLMALONYL(CARBADETHIA)-COENZYME A, ... | | Authors: | Evans, P.R, Thomae, N.H. | | Deposit date: | 1998-08-03 | | Release date: | 1998-08-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Stabilization of radical intermediates by an active-site tyrosine residue in methylmalonyl-CoA mutase.
Biochemistry, 37, 1998
|
|
1E1C
 
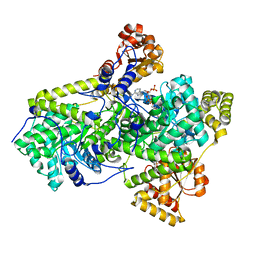 | | METHYLMALONYL-COA MUTASE H244A Mutant | | Descriptor: | COBALAMIN, DESULFO-COENZYME A, METHYLMALONYL-COA MUTASE ALPHA CHAIN, ... | | Authors: | Evans, P.R, Thoma, N.H. | | Deposit date: | 2000-04-30 | | Release date: | 2000-05-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Protection of Radical Intermediates at the Active Site of Adenosylcobalamin-Dependent Methylmalonyl-Coa Mutase
Biochemistry, 39, 2000
|
|
8R1G
 
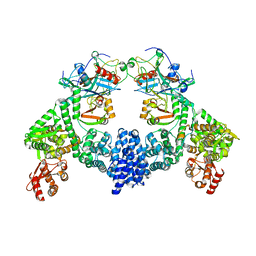 | | Dimeric ternary structure of E6AP-E6-p53 | | Descriptor: | Cellular tumor antigen p53, Ubiquitin-like protein SMT3,Protein E6, Ubiquitin-protein ligase E3A, ... | | Authors: | Sandate, C.R, Chakrabory, D, Kater, L, Kempf, G, Thoma, N.H. | | Deposit date: | 2023-11-01 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Monomeric E6AP-E6-p53 ternary complex
To Be Published
|
|
8R1F
 
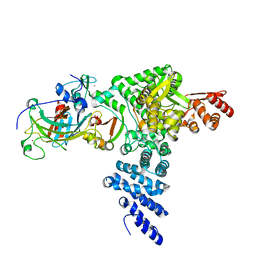 | | Monomeric E6AP-E6-p53 ternary complex | | Descriptor: | Cellular tumor antigen p53, Ubiquitin-like protein SMT3,Protein E6, Ubiquitin-protein ligase E3A, ... | | Authors: | Sandate, C.R, Chakraborty, D, Kater, L, Kempf, G, Thoma, N.H. | | Deposit date: | 2023-11-01 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Monomeric E6AP-E6-p53 ternary complex
To Be Published
|
|
8OSK
 
 | | Cryo-EM structure of CLOCK-BMAL1 bound to a nucleosomal E-box at position SHL+5.8 (composite map) | | Descriptor: | Basic helix-loop-helix ARNT-like protein 1, Circadian locomoter output cycles protein kaput, DNA (124-MER), ... | | Authors: | Stoos, L, Michael, A.K, Kempf, G, Cavadini, S, Thoma, N.H. | | Deposit date: | 2023-04-19 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
8OSJ
 
 | | Cryo-EM structure of CLOCK-BMAL1 bound to a nucleosomal E-box at position SHL-6.2 (DNA conformation 1) | | Descriptor: | Basic helix-loop-helix ARNT-like protein 1, Circadian locomoter output cycles protein kaput, DNA (124-MER), ... | | Authors: | Michael, A.K, Stoos, L, Kempf, G, Cavadini, S, Thoma, N.H. | | Deposit date: | 2023-04-19 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
8P83
 
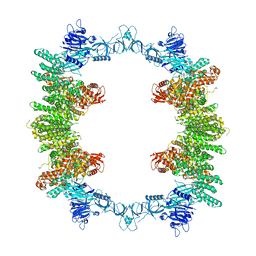 | | Cryo-EM structure of full-length human UBR5 (homotetramer) | | Descriptor: | E3 ubiquitin-protein ligase UBR5 | | Authors: | Aguirre, J.D, Kater, L, Kempf, G, Cavadini, S, Thoma, N.H. | | Deposit date: | 2023-05-31 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | UBR5 forms ligand-dependent complexes on chromatin to regulate nuclear hormone receptor stability.
Mol.Cell, 83, 2023
|
|
5NVR
 
 | |
5NW5
 
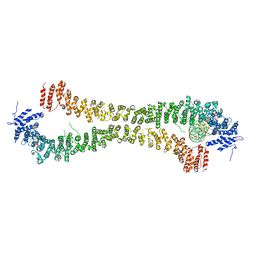 | | Crystal structure of the Rif1 N-terminal domain (RIF1-NTD) from Saccharomyces cerevisiae in complex with DNA | | Descriptor: | DNA (30-MER), DNA (60-MER), Telomere length regulator protein RIF1 | | Authors: | Bunker, R.D, Reinert, J.K, Shi, T, Thoma, N.H. | | Deposit date: | 2017-05-05 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (6.502 Å) | | Cite: | Rif1 maintains telomeres and mediates DNA repair by encasing DNA ends.
Nat. Struct. Mol. Biol., 24, 2017
|
|
4WSN
 
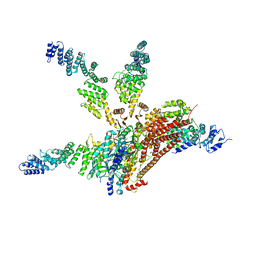 | | Crystal structure of the COP9 signalosome, a P1 crystal form | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Bunker, R.D, Lingaraju, G.M, Thoma, N.H. | | Deposit date: | 2014-10-28 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Cullin-RING ubiquitin E3 ligase regulation by the COP9 signalosome.
Nature, 531, 2016
|
|
6GVW
 
 | | Crystal structure of the BRCA1-A complex | | Descriptor: | BRCA1-A complex subunit Abraxas 1, BRCA1-A complex subunit RAP80, BRISC and BRCA1-A complex member 1, ... | | Authors: | Bunker, R.D, Rabl, J, Thoma, N.H. | | Deposit date: | 2018-06-21 | | Release date: | 2019-07-10 | | Last modified: | 2019-08-21 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Structural Basis of BRCC36 Function in DNA Repair and Immune Regulation.
Mol.Cell, 75, 2019
|
|
6H0F
 
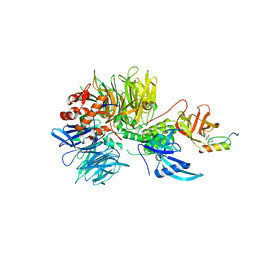 | | Structure of DDB1-CRBN-pomalidomide complex bound to IKZF1(ZF2) | | Descriptor: | DNA damage-binding protein 1,DNA damage-binding protein 1,DNA damage-binding protein 1,DNA damage-binding protein 1, DNA-binding protein Ikaros, Protein cereblon, ... | | Authors: | Petzold, G, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2018-07-09 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Defining the human C2H2 zinc finger degrome targeted by thalidomide analogs through CRBN.
Science, 362, 2018
|
|
6H0G
 
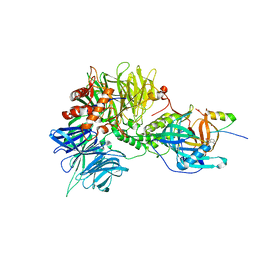 | | Structure of the DDB1-CRBN-pomalidomide complex bound to ZNF692(ZF4) | | Descriptor: | DNA damage-binding protein 1,DNA damage-binding protein 1,DNA damage-binding protein 1,DDB1 (DNA damage binding protein 1),DNA damage-binding protein 1,DNA damage-binding protein 1,DNA damage-binding protein 1, Protein cereblon, S-Pomalidomide, ... | | Authors: | Bunker, R.D, Petzold, G, Thoma, N.H. | | Deposit date: | 2018-07-09 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (4.25 Å) | | Cite: | Defining the human C2H2 zinc finger degrome targeted by thalidomide analogs through CRBN.
Science, 362, 2018
|
|
6H3C
 
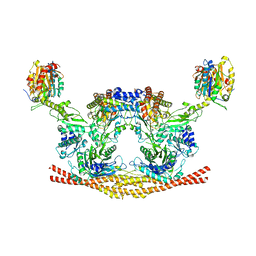 | | Cryo-EM structure of the BRISC complex bound to SHMT2 | | Descriptor: | BRISC and BRCA1-A complex member 1, BRISC and BRCA1-A complex member 2, BRISC complex subunit Abraxas 2, ... | | Authors: | Bunker, R.D, Rabl, J, Thoma, N.H. | | Deposit date: | 2018-07-18 | | Release date: | 2019-07-10 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Basis of BRCC36 Function in DNA Repair and Immune Regulation.
Mol.Cell, 75, 2019
|
|
8AJO
 
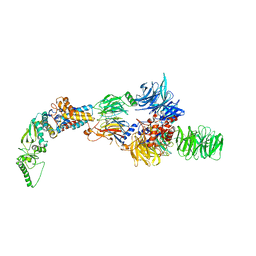 | | Negative-stain electron microscopy structure of DDB1-DCAF12-CCT5 | | Descriptor: | DDB1- and CUL4-associated factor 12, DNA damage-binding protein 1, T-complex protein 1 subunit epsilon | | Authors: | Pla-Prats, C, Cavadini, S, Kempf, G, Thoma, N.H. | | Deposit date: | 2022-07-28 | | Release date: | 2022-11-09 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (30.6 Å) | | Cite: | Recognition of the CCT5 di-Glu degron by CRL4 DCAF12 is dependent on TRiC assembly.
Embo J., 42, 2023
|
|
8AJM
 
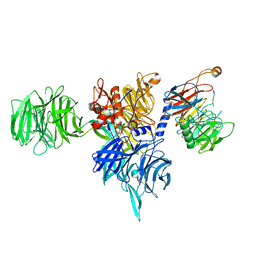 | | Structure of human DDB1-DCAF12 in complex with the C-terminus of CCT5 | | Descriptor: | DDB1- and CUL4-associated factor 12, DNA damage-binding protein 1, T-complex protein 1 subunit epsilon | | Authors: | Pla-Prats, C, Cavadini, S, Kempf, G, Thoma, N.H. | | Deposit date: | 2022-07-28 | | Release date: | 2022-11-09 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Recognition of the CCT5 di-Glu degron by CRL4 DCAF12 is dependent on TRiC assembly.
Embo J., 42, 2023
|
|
8AJN
 
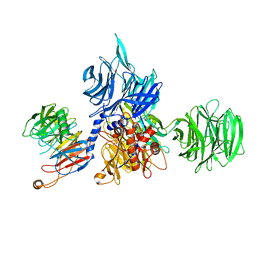 | | Structure of the human DDB1-DCAF12 complex | | Descriptor: | DDB1- and CUL4-associated factor 12, DNA damage-binding protein 1 | | Authors: | Pla-Prats, C, Cavadini, S, Kempf, G, Thoma, N.H. | | Deposit date: | 2022-07-28 | | Release date: | 2022-11-09 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Recognition of the CCT5 di-Glu degron by CRL4 DCAF12 is dependent on TRiC assembly.
Embo J., 42, 2023
|
|
7OKQ
 
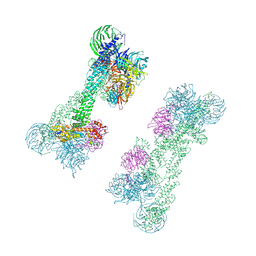 | | Cryo-EM Structure of the DDB1-DCAF1-CUL4A-RBX1 Complex | | Descriptor: | Cullin-4A, DDB1- and CUL4-associated factor 1, DNA damage-binding protein 1, ... | | Authors: | Mohamed, W.I, Schenk, A.D, Kempf, G, Cavadini, S, Thoma, N.H. | | Deposit date: | 2021-05-18 | | Release date: | 2021-10-13 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | The CRL4 DCAF1 cullin-RING ubiquitin ligase is activated following a switch in oligomerization state.
Embo J., 40, 2021
|
|
1LTX
 
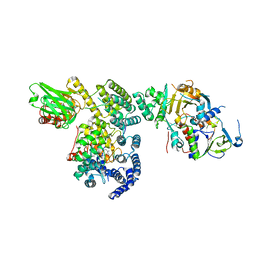 | | Structure of Rab Escort Protein-1 in complex with Rab geranylgeranyl transferase and isoprenoid | | Descriptor: | AAAA, CHLORIDE ION, FARNESYL, ... | | Authors: | Pylypenko, O, Rak, A, Reents, R, Niculae, A, Thoma, N.H, Waldmann, H, Schlichting, I, Goody, R.S, Alexandrov, K. | | Deposit date: | 2002-05-21 | | Release date: | 2003-05-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of Rab Escort Protein-1 in Complex with Rab Geranylgeranyltransferase
Mol.Cell, 11, 2003
|
|
4YM6
 
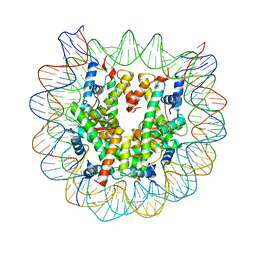 | | Crystal structure of the human nucleosome containing 6-4PP (outside) | | Descriptor: | 145-MER DNA, Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Osakabe, A, Tachiwana, H, Kagawa, W, Horikoshi, N, Matsumoto, S, Hasegawa, M, Matsumoto, N, Toga, T, Yamamoto, J, Hanaoka, F, Thoma, N.H, Sugasawa, K, Iwai, S, Kurumizaka, H. | | Deposit date: | 2015-03-06 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.514 Å) | | Cite: | Structural basis of pyrimidine-pyrimidone (6-4) photoproduct recognition by UV-DDB in the nucleosome
Sci Rep, 5, 2015
|
|
4YM5
 
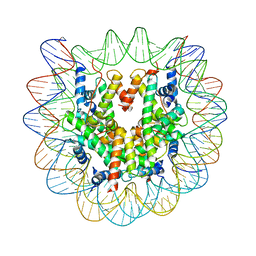 | | Crystal structure of the human nucleosome containing 6-4PP (inside) | | Descriptor: | 144 mer-DNA, 144-mer DNA, Histone H2A type 1-B/E, ... | | Authors: | Osakabe, A, Tachiwana, H, Kagawa, W, Horikoshi, N, Matsumoto, S, Hasegawa, M, Matsumoto, N, Toga, T, Yamamoto, J, Hanaoka, F, Thoma, N.H, Sugasawa, K, Iwai, S, Kurumizaka, H. | | Deposit date: | 2015-03-06 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.005 Å) | | Cite: | Structural basis of pyrimidine-pyrimidone (6-4) photoproduct recognition by UV-DDB in the nucleosome
Sci Rep, 5, 2015
|
|
4A0B
 
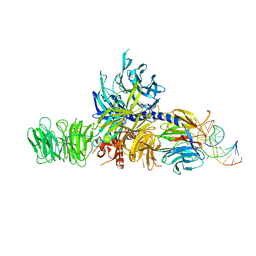 | | Structure of hsDDB1-drDDB2 bound to a 16 bp CPD-duplex (pyrimidine at D-1 position) at 3.8 A resolution (CPD 4) | | Descriptor: | 5'-D(*CP*CP*TP*GP*CP*TP*CP*CP*TP*TP*TP*CP*AP*CP*CP*C)-3', 5'-D(*DGP*GP*TP*GP*AP*AP*AP*(TTD)P*AP*GP*CP*AP*GP*DGP)-3', DNA DAMAGE-BINDING PROTEIN 1, ... | | Authors: | Scrima, A, Fischer, E.S, Iwai, S, Gut, H, Thoma, N.H. | | Deposit date: | 2011-09-08 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation
Cell(Cambridge,Mass.), 147, 2011
|
|
4A0L
 
 | | Structure of DDB1-DDB2-CUL4B-RBX1 bound to a 12 bp abasic site containing DNA-duplex | | Descriptor: | 12 BP DNA DUPLEX, 12 BP THF CONTAINING DNA DUPLEX, CULLIN-4B, ... | | Authors: | Fischer, E.S, Scrima, A, Gut, H, Thoma, N.H. | | Deposit date: | 2011-09-09 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (7.4 Å) | | Cite: | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation.
Cell(Cambridge,Mass.), 147, 2011
|
|
4A0A
 
 | | Structure of hsDDB1-drDDB2 bound to a 16 bp CPD-duplex (pyrimidine at D-1 position) at 3.6 A resolution (CPD 3) | | Descriptor: | 5'-D(*CP*CP*TP*GP*CP*TP*CP*CP*TP*TP*TP*CP*AP*CP*CP*C)-3', 5'-D(*GP*GP*TP*GP*AP*AP*AP*(TTD)P*AP*GP*CP*AP*GP*DGP)-3', CALCIUM ION, ... | | Authors: | Scrima, A, Fischer, E.S, Iwai, S, Gut, H, Thoma, N.H. | | Deposit date: | 2011-09-08 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation
Cell(Cambridge,Mass.), 147, 2011
|
|
