8S9T
 
 | |
8S9U
 
 | |
8S9V
 
 | |
8S9X
 
 | |
8EW5
 
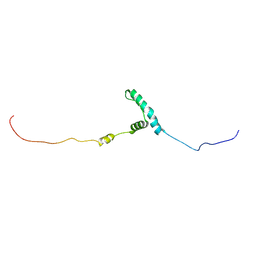 | | The structure of flightin within myosin thick filaments from Bombus ignitus flight muscle | | Descriptor: | Flightin | | Authors: | Li, J, Rahmani, H, Abbasi Yeganeh, F, Rastegarpouyani, H, Taylor, D.W, Wood, N.B, Previs, M.J, Iwamoto, H, Taylor, K.A. | | Deposit date: | 2022-10-21 | | Release date: | 2023-01-04 | | Last modified: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structure of the Flight Muscle Thick Filament from the Bumble Bee, Bombus ignitus , at 6 angstrom Resolution.
Int J Mol Sci, 24, 2022
|
|
8SYF
 
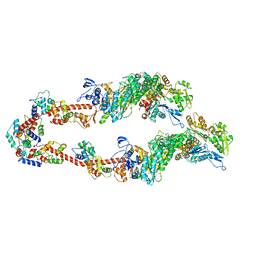 | | Homology model of Acto-HMM complex in ADP-state. Chicken smooth muscle HMM and chicken pectoralis actin | | Descriptor: | Actin, alpha skeletal muscle, Myosin light polypeptide 6, ... | | Authors: | Hojjatian, A, Taylor, D.W, Daneshparvar, N, Trybus, K.M, Taylor, K.A. | | Deposit date: | 2023-05-25 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (19 Å) | | Cite: | Double-headed binding of myosin II to F-actin shows the effect of strain on head structure.
J.Struct.Biol., 215, 2023
|
|
8TL0
 
 | |
6MDR
 
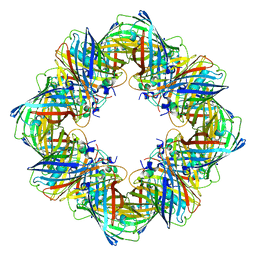 | | Cryo-EM structure of the Ceru+32/GFP-17 protomer | | Descriptor: | Ceru+32, GFP-17 | | Authors: | Simon, A.J, Zhou, Y, Ramasubramani, V, Glaser, J, Pothukuchy, A, Golihar, J, Gerberich, J.C, Leggere, J.C, Morrow, B.R, Jung, C, Glotzer, S.C, Taylor, D.W, Ellington, A.D. | | Deposit date: | 2018-09-05 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Supercharging enables organized assembly of synthetic biomolecules.
Nat Chem, 11, 2019
|
|
6N8K
 
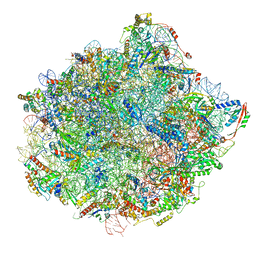 | | Cryo-EM structure of early cytoplasmic-immediate (ECI) pre-60S ribosomal subunit | | Descriptor: | 5.8S rRNA, 5S rRNA, 60S ribosomal export protein NMD3, ... | | Authors: | Zhou, Y, Musalgaonkar, S, Johnson, A.W, Taylor, D.W. | | Deposit date: | 2018-11-29 | | Release date: | 2019-03-13 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Tightly-orchestrated rearrangements govern catalytic center assembly of the ribosome.
Nat Commun, 10, 2019
|
|
1SJJ
 
 | |
6N8O
 
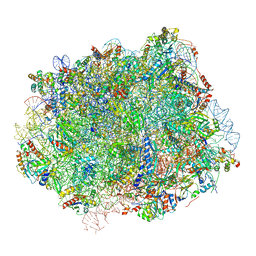 | | Cryo-EM structure of Rpl10-inserted (RI) pre-60S ribosomal subunit | | Descriptor: | 5.8S rRNA, 5S rRNA, 60S ribosomal export protein NMD3, ... | | Authors: | Zhou, Y, Musalgaonkar, S, Johnson, A.W, Taylor, D.W. | | Deposit date: | 2018-11-29 | | Release date: | 2019-03-13 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Tightly-orchestrated rearrangements govern catalytic center assembly of the ribosome.
Nat Commun, 10, 2019
|
|
6N8J
 
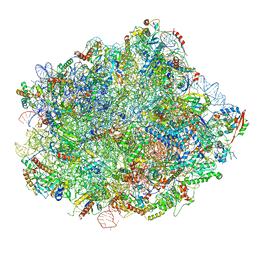 | | Cryo-EM structure of late nuclear (LN) pre-60S ribosomal subunit | | Descriptor: | 5.8S rRNA, 5S rRNA, 60S ribosomal protein L11-A, ... | | Authors: | Zhou, Y, Musalgaonkar, S, Johnson, A.W, Taylor, D.W. | | Deposit date: | 2018-11-29 | | Release date: | 2019-03-13 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Tightly-orchestrated rearrangements govern catalytic center assembly of the ribosome.
Nat Commun, 10, 2019
|
|
7S4X
 
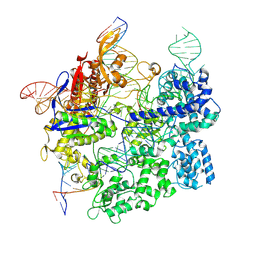 | | Cas9:gRNA in complex with 18-20MM DNA, 1 minute time-point, kinked active conformation | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, MAGNESIUM ION, NTS, ... | | Authors: | Bravo, J.P.K, Taylor, D.W, Liu, M.S, Johnson, K.A. | | Deposit date: | 2021-09-09 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural basis for mismatch surveillance by CRISPR-Cas9.
Nature, 603, 2022
|
|
7S4U
 
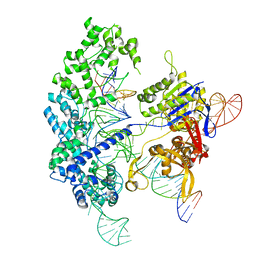 | | Cryo-EM structure of Cas9 in complex with 12-14MM DNA substrate, 5 minute time-point | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, Non-target strand, Target strand, ... | | Authors: | Bravo, J.P.K, Taylor, D.W, Liu, M.S, Johnson, K.A. | | Deposit date: | 2021-09-09 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Structural basis for mismatch surveillance by CRISPR-Cas9.
Nature, 603, 2022
|
|
7SBB
 
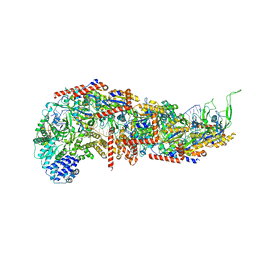 | |
7SBA
 
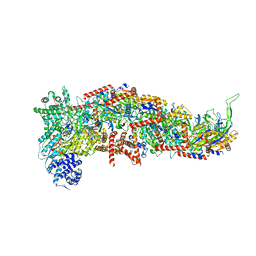 | |
7S9V
 
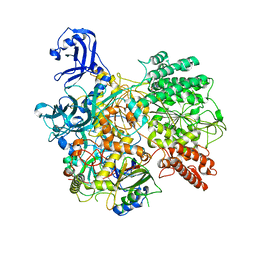 | | DrmAB:ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DrmA, DrmB | | Authors: | Bravo, J.P.K, Taylor, D.W, Brouns, S.J.J, Aparicio-Maldonado, C. | | Deposit date: | 2021-09-21 | | Release date: | 2022-06-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for broad anti-phage immunity by DISARM.
Nat Commun, 13, 2022
|
|
7S9W
 
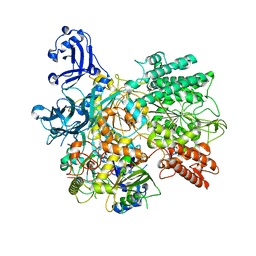 | | Structure of DrmAB:ADP:DNA complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*T)-3'), DrmA, ... | | Authors: | Bravo, J.P.K, Taylor, D.W, Brounds, S.J.J, Aparicio-Maldonado, C. | | Deposit date: | 2021-09-21 | | Release date: | 2022-06-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for broad anti-phage immunity by DISARM.
Nat Commun, 13, 2022
|
|
7UG6
 
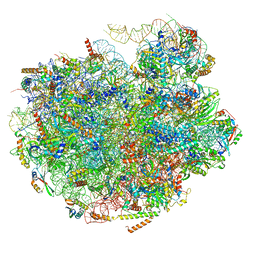 | | Cryo-EM structure of pre-60S ribosomal subunit, unmethylated G2922 | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Yelland, J.N, Bravo, J.P.K, Black, J.J.B, Taylor, D.W, Johnson, A.W. | | Deposit date: | 2022-03-24 | | Release date: | 2022-12-21 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | A single 2'-O-methylation of ribosomal RNA gates assembly of a functional ribosome.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8D4B
 
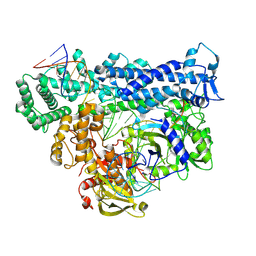 | | Structure of Cas12a2 ternary complex | | Descriptor: | OrfB_Zn_ribbon domain-containing protein, RNA (28-MER), RNA (41-MER) | | Authors: | Bravo, J.P.K, Taylor, D.W. | | Deposit date: | 2022-06-01 | | Release date: | 2023-01-18 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | RNA targeting unleashes indiscriminate nuclease activity of CRISPR-Cas12a2.
Nature, 613, 2023
|
|
8D4A
 
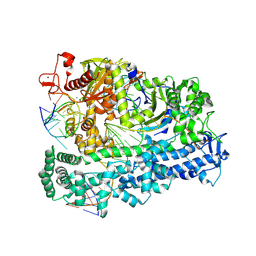 | | Cas12a2 quaternary complex | | Descriptor: | DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), MAGNESIUM ION, ... | | Authors: | Bravo, J.P.K, Taylor, D.W. | | Deposit date: | 2022-06-01 | | Release date: | 2023-01-18 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | RNA targeting unleashes indiscriminate nuclease activity of CRISPR-Cas12a2.
Nature, 613, 2023
|
|
8D49
 
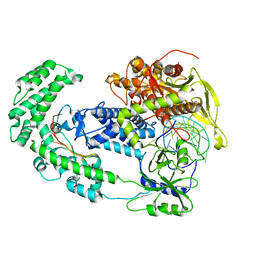 | | Structure of Cas12a2 binary complex | | Descriptor: | OrfB_Zn_ribbon domain-containing protein, RNA (26-MER) | | Authors: | Bravo, J.P.K, Taylor, D.W. | | Deposit date: | 2022-06-01 | | Release date: | 2023-01-18 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | RNA targeting unleashes indiscriminate nuclease activity of CRISPR-Cas12a2.
Nature, 613, 2023
|
|
8DFO
 
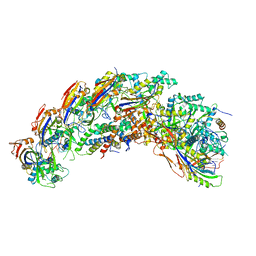 | | type I-C Cascade bound to AcrIC4 | | Descriptor: | AcrIC4, CRISPR-associated protein, CT1133 family, ... | | Authors: | O'Brien, R.E, Bravo, J.P.K, Ramos, D, Hibshman, G.N, Wright, J.T, Taylor, D.W. | | Deposit date: | 2022-06-22 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural snapshots of R-loop formation by a type I-C CRISPR Cascade.
Mol.Cell, 83, 2023
|
|
8DEX
 
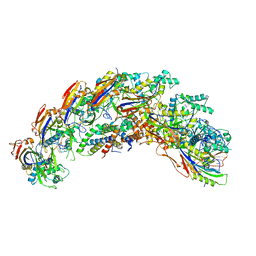 | | type I-C Cascade | | Descriptor: | CRISPR-associated protein, CT1133 family, TM1801 family, ... | | Authors: | O'Brien, R.E, Bravo, J.P.K, Ramos, D, Hibshman, G.N, Wright, J.T, Taylor, D.W. | | Deposit date: | 2022-06-21 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural snapshots of R-loop formation by a type I-C CRISPR Cascade.
Mol.Cell, 83, 2023
|
|
8DEJ
 
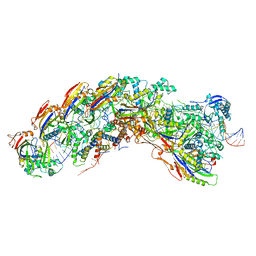 | | D. vulgaris type I-C Cascade bound to dsDNA target | | Descriptor: | CRISPR-associated protein, CT1133 family, TM1801 family, ... | | Authors: | O'Brien, R.E, Bravo, J.P.K, Ramos, D, Hibshman, G.N, Wright, J.T, Taylor, D.W. | | Deposit date: | 2022-06-20 | | Release date: | 2023-02-22 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural snapshots of R-loop formation by a type I-C CRISPR Cascade.
Mol.Cell, 83, 2023
|
|
