1CYU
 
 | | SOLUTION NMR STRUCTURE OF RECOMBINANT HUMAN CYSTATIN A UNDER THE CONDITION OF PH 3.8 AND 310K | | Descriptor: | CYSTATIN A | | Authors: | Tate, S, Tate, N.U, Ushioda, T, Samejima, T, Kainosho, M. | | Deposit date: | 1995-08-24 | | Release date: | 1995-12-07 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a human cystatin A variant, cystatin A2-98 M65L, by NMR spectroscopy. A possible role of the interactions between the N- and C-termini to maintain the inhibitory active form of cystatin A.
Biochemistry, 34, 1995
|
|
1CYV
 
 | | SOLUTION NMR STRUCTURE OF RECOMBINANT HUMAN CYSTATIN A UNDER THE CONDITION OF PH 3.8 AND 310K | | Descriptor: | CYSTATIN A | | Authors: | Tate, S, Tate, N.U, Ushioda, T, Samejima, T, Kainosho, M. | | Deposit date: | 1995-08-24 | | Release date: | 1995-12-07 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a human cystatin A variant, cystatin A2-98 M65L, by NMR spectroscopy. A possible role of the interactions between the N- and C-termini to maintain the inhibitory active form of cystatin A.
Biochemistry, 34, 1995
|
|
1BHU
 
 | | THE 3D STRUCTURE OF THE STREPTOMYCES METALLOPROTEINASE INHIBITOR, SMPI, ISOLATED FROM STREPTOMYCES NIGRESCENS TK-23, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | METALLOPROTEINASE INHIBITOR | | Authors: | Tate, S, Ohno, A, Seeram, S.S, Hiraga, K, Oda, K, Kainosho, M. | | Deposit date: | 1998-06-10 | | Release date: | 1999-01-06 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the Streptomyces metalloproteinase inhibitor, SMPI, isolated from Streptomyces nigrescens TK-23: another example of an ancestral beta gamma-crystallin precursor structure.
J.Mol.Biol., 282, 1998
|
|
5GPH
 
 | |
1YXJ
 
 | | Crystal structure of human lectin-like oxidized low-density lipoprotein receptor 1 (LOX-1) at low pH | | Descriptor: | 1,2-ETHANEDIOL, oxidised low density lipoprotein (lectin-like) receptor 1 | | Authors: | Ohki, I, Ishigaki, T, Oyama, T, Matsunaga, S, Xie, Q, Ohnishi-Kameyama, M, Murata, T, Tsuchiya, D, Machida, S, Morikawa, K, Tate, S. | | Deposit date: | 2005-02-22 | | Release date: | 2005-06-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of human lectin-like, oxidized low-density lipoprotein receptor 1 ligand binding domain and its ligand recognition mode to OxLDL.
Structure, 13, 2005
|
|
1YXK
 
 | | Crystal structure of human lectin-like oxidized low-density lipoprotein receptor 1 (LOX-1) disulfide-linked dimer | | Descriptor: | oxidised low density lipoprotein (lectin-like) receptor 1 | | Authors: | Ohki, I, Ishigaki, T, Oyama, T, Matsunaga, S, Xie, Q, Ohnishi-Kameyama, M, Murata, T, Tsuchiya, D, Machida, S, Morikawa, K, Tate, S. | | Deposit date: | 2005-02-22 | | Release date: | 2005-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human lectin-like, oxidized low-density lipoprotein receptor 1 ligand binding domain and its ligand recognition mode to OxLDL.
Structure, 13, 2005
|
|
3VLG
 
 | | Crystal structure of the W150A mutant LOX-1 CTLD showing impaired OxLDL binding | | Descriptor: | Oxidized low-density lipoprotein receptor 1 | | Authors: | Nakano, S, Sugihara, M, Yamada, R, Katayanagi, K, Tate, S. | | Deposit date: | 2011-12-01 | | Release date: | 2012-04-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural implication for the impaired binding of W150A mutant LOX-1 to oxidized low density lipoprotein, OxLDL
Biochim.Biophys.Acta, 1824, 2012
|
|
2CV5
 
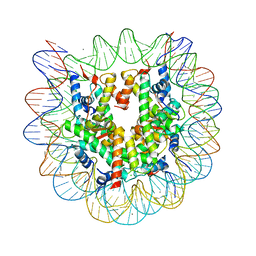 | | Crystal structure of human nucleosome core particle | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A.a, ... | | Authors: | Tsunaka, Y, Kajimura, N, Tate, S, Morikawa, K. | | Deposit date: | 2005-05-31 | | Release date: | 2005-06-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Alteration of the nucleosomal DNA path in the crystal structure of a human nucleosome core particle
Nucleic Acids Res., 33, 2005
|
|
1WXL
 
 | | Solution Structure of the HMG-box domain in the SSRP1 subunit of FACT | | Descriptor: | Single-strand recognition protein | | Authors: | Kasai, N, Tsunaka, Y, Ohki, I, Hirose, S, Morikawa, K, Tate, S. | | Deposit date: | 2005-01-26 | | Release date: | 2005-08-16 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the HMG-box domain in the SSRP1 subunit of FACT
J.Biomol.Nmr, 32, 2005
|
|
1DUF
 
 | | THE NMR STRUCTURE OF DNA DODECAMER DETERMINED IN AQUEOUS DILUTE LIQUID CRYSTALLINE PHASE | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Tjandra, N, Tate, S, Ono, A, Kainosho, M, Bax, A. | | Deposit date: | 2000-01-17 | | Release date: | 2000-07-14 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The NMR Structure of a DNA Dodecamer in an Aqueous Dilute Liquid Crystalline Phase
J.Am.Chem.Soc., 122, 2000
|
|
1GD4
 
 | | SOLUTION STRUCTURE OF P25S CYSTATIN A | | Descriptor: | CYSTATIN A | | Authors: | Shimba, N, Kariya, E, Tate, S, Kaji, H, Kainosho, M. | | Deposit date: | 2000-09-08 | | Release date: | 2001-09-08 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural comparison between wild-type and P25S human cystatin A by NMR spectroscopy. Does this mutation affect the a-helix conformation ?
J.STRUCT.FUNCT.GENOM., 1, 2000
|
|
1GD3
 
 | | refined solution structure of human cystatin A | | Descriptor: | CYSTATIN A | | Authors: | Shimba, N, Kariya, E, Tate, S, Kaji, H, Kainosho, M. | | Deposit date: | 2000-09-08 | | Release date: | 2001-09-08 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural comparison between wild-type and P25S human cystatin A by NMR spectroscopy. Does this mutation affect the a-helix
conformation ?
J.STRUCT.FUNCT.GENOM., 1, 2000
|
|
2RUC
 
 | |
2RUQ
 
 | | solution structure of human Pin1 PPIase mutant C113A | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Jing, W, Tochio, N, Tate, S. | | Deposit date: | 2015-01-20 | | Release date: | 2016-01-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Allosteric Breakage of the Hydrogen Bond within the Dual-Histidine Motif in the Active Site of Human Pin1 PPIase
Biochemistry, 54, 2015
|
|
2RUD
 
 | |
2RUR
 
 | | Solution structure of Human Pin1 PPIase C113S mutant | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Jing, W, Tochio, N, Tate, S. | | Deposit date: | 2015-01-20 | | Release date: | 2016-01-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Allosteric Breakage of the Hydrogen Bond within the Dual-Histidine Motif in the Active Site of Human Pin1 PPIase
Biochemistry, 54, 2015
|
|
1J3D
 
 | |
1J3C
 
 | |
1J3X
 
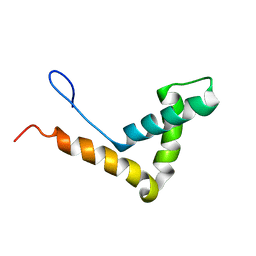 | |
2RRL
 
 | | Solution structure of the C-terminal domain of the FliK | | Descriptor: | Flagellar hook-length control protein | | Authors: | Mizuno, S, Tate, S, Kobayashi, N, Amida, H. | | Deposit date: | 2011-01-02 | | Release date: | 2011-05-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of FliK, the trigger for the switch of substrate specificity in the flagellar type III secretion apparatus
J.Mol.Biol., 409, 2011
|
|
2RTU
 
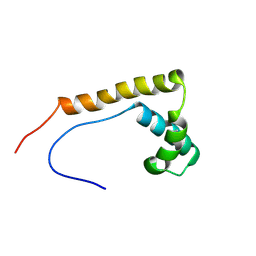 | | Solution structure of oxidized human HMGB1 A box | | Descriptor: | High mobility group protein B1 | | Authors: | Jing, W, Tochio, N, Tate, S. | | Deposit date: | 2013-09-12 | | Release date: | 2014-03-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Redox-sensitive structural change in the A-domain of HMGB1 and its implication for the binding to cisplatin modified DNA.
Biochem.Biophys.Res.Commun., 441, 2013
|
|
5XIR
 
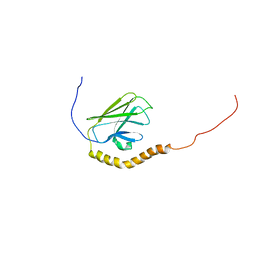 | |
5XI9
 
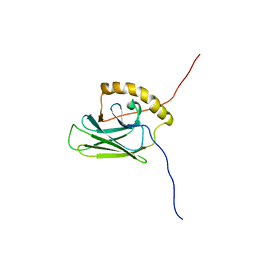 | |
1ERA
 
 | |
1FRA
 
 | |
