3LQB
 
 | | Crystal structure of the hatching enzyme ZHE1 from the zebrafish Danio rerio | | Descriptor: | 1,2-ETHANEDIOL, LOC792177 protein, SULFATE ION, ... | | Authors: | Tanokura, M, Okada, A, Nagata, K, Yasumasu, S, Ohtsuka, J, Iuchi, I. | | Deposit date: | 2010-02-08 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structure of zebrafish hatching enzyme 1 from the zebrafish Danio rerio
J.Mol.Biol., 402, 2010
|
|
3AEV
 
 | | Crystal structure of a/eIF2alpha-aDim2p-rRNA complex from Pyrococcus horikoshii OT3 | | Descriptor: | Putative uncharacterized protein PH1566, RNA (5'-R(*GP*GP*AP*UP*CP*AP*CP*CP*UP*CP*C)-3'), Translation initiation factor 2 subunit alpha | | Authors: | Tanokura, M, Jia, M.Z, Nagata, K. | | Deposit date: | 2010-02-10 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | An archaeal Dim2-like protein, aDim2p, forms a ternary complex with a/eIF2 alpha and the 3' end fragment of 16S rRNA
J.Mol.Biol., 398, 2010
|
|
2E3U
 
 | |
4WFJ
 
 | | Crystal structure of PET-degrading cutinase Cut190 S226P mutant in Ca(2+)-bound state at 1.75 angstrom resolution | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cutinase | | Authors: | Miyakawa, T, Mizushima, H, Ohtsuka, J, Oda, M, Kawai, F, Tanokura, M. | | Deposit date: | 2014-09-15 | | Release date: | 2014-12-24 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for the Ca(2+)-enhanced thermostability and activity of PET-degrading cutinase-like enzyme from Saccharomonospora viridis AHK190.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
4WFK
 
 | | Crystal structure of PET-degrading cutinase Cut190 S226P mutant in Ca(2+)-bound state at 2.35 angstrom resolution | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cutinase | | Authors: | Miyakawa, T, Mizushima, H, Ohtsuka, J, Oda, M, Kawai, F, Tanokura, M. | | Deposit date: | 2014-09-15 | | Release date: | 2014-12-24 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for the Ca(2+)-enhanced thermostability and activity of PET-degrading cutinase-like enzyme from Saccharomonospora viridis AHK190.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
4O0L
 
 | |
2MK4
 
 | | Solution structure of ORF2 | | Descriptor: | Open reading frame 2 | | Authors: | Miyakawa, T, Kobayashi, H, Tashiro, M, Yamanaka, H, Tanokura, M. | | Deposit date: | 2014-01-24 | | Release date: | 2015-03-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Action of the External Chaperone for a Propeptide-deficient Serine Protease from Aeromonas sobria.
J.Biol.Chem., 290, 2015
|
|
5TLC
 
 | |
5WVU
 
 | | Crystal structure of carboxypeptidase from Thermus thermophilus | | Descriptor: | GLYCEROL, Thermostable carboxypeptidase 1, ZINC ION | | Authors: | Okai, M, Nagata, K, Tanokura, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2016-12-29 | | Release date: | 2017-02-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insight into the transition between the open and closed conformations of Thermus thermophilus carboxypeptidase.
Biochem. Biophys. Res. Commun., 484, 2017
|
|
1DET
 
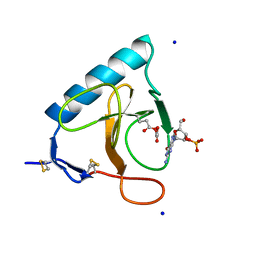 | | RIBONUCLEASE T1 CARBOXYMETHYLATED AT GLU 58 IN COMPLEX WITH 2'GMP | | Descriptor: | GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE T1, SODIUM ION | | Authors: | Ishikawa, K, Suzuki, E, Tanokura, M, Takahashi, K. | | Deposit date: | 1996-02-20 | | Release date: | 1996-07-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of ribonuclease T1 carboxymethylated at Glu58 in complex with 2'-GMP.
Biochemistry, 35, 1996
|
|
1IYY
 
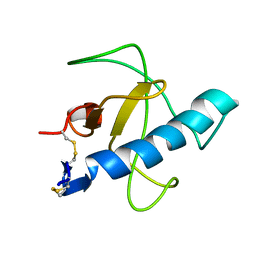 | | NMR STRUCTURE OF Gln25-RIBONUCLEASE T1, 24 STRUCTURES | | Descriptor: | RIBONUCLEASE T1 | | Authors: | Hatano, K, Kojima, M, Suzuki, E, Tanokura, M, Takahashi, K. | | Deposit date: | 2002-09-12 | | Release date: | 2003-10-07 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Determination of the NMR structure of Gln25-ribonuclease T1.
Biol. Chem., 384, 2003
|
|
4TMC
 
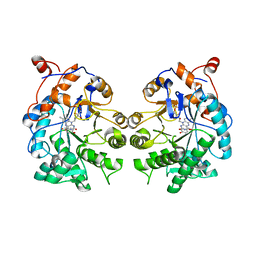 | | CRYSTAL STRUCTURE of OLD YELLOW ENZYME from CANDIDA MACEDONIENSIS AKU4588 COMPLEXED with P-HYDROXYBENZALDEHYDE | | Descriptor: | FLAVIN MONONUCLEOTIDE, Old yellow enzyme, P-HYDROXYBENZALDEHYDE | | Authors: | Horita, S, Kataoka, M, Kitamura, N, Nakagawa, T, Miyakawa, T, Ohtsuka, J, Nagata, K, Shimizu, S, Tanokura, M. | | Deposit date: | 2014-05-31 | | Release date: | 2015-02-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Engineered Old Yellow Enzyme that Enables Efficient Synthesis of (4R,6R)-Actinol in a One-Pot Reduction System
Chembiochem, 16, 2015
|
|
4TMB
 
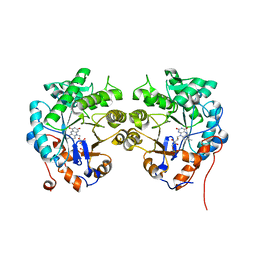 | | CRYSTAL STRUCTURE of OLD YELLOW ENZYME from CANDIDA MACEDONIENSIS AKU4588 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Old yellow enzyme | | Authors: | Horita, S, Kataoka, M, Kitamura, N, Nakagawa, T, Miyakawa, T, Ohtsuka, J, Nagata, K, Shimizu, S, Tanokura, M. | | Deposit date: | 2014-05-31 | | Release date: | 2015-02-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Engineered Old Yellow Enzyme that Enables Efficient Synthesis of (4R,6R)-Actinol in a One-Pot Reduction System
Chembiochem, 16, 2015
|
|
4QLX
 
 | | Crystal structure of CLA-ER with product binding | | Descriptor: | 10-oxooctadecanoic acid, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Hou, F, Miyakawa, T, Tanokura, M. | | Deposit date: | 2014-06-13 | | Release date: | 2015-02-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and reaction mechanism of a novel enone reductase.
Febs J., 282, 2015
|
|
4QLY
 
 | | Crystal structure of CLA-ER, a novel enone reductase catalyzing a key step of a gut-bacterial fatty acid saturation metabolism, biohydrogenation | | Descriptor: | Enone reductase CLA-ER, FLAVIN MONONUCLEOTIDE | | Authors: | Hou, F, Miyakawa, T, Tanokura, M. | | Deposit date: | 2014-06-13 | | Release date: | 2015-02-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Structure and reaction mechanism of a novel enone reductase.
Febs J., 282, 2015
|
|
1EQK
 
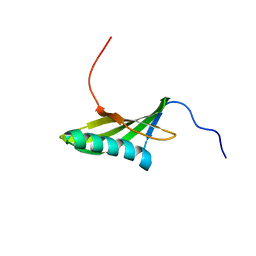 | | SOLUTION STRUCTURE OF ORYZACYSTATIN-I, A CYSTEINE PROTEINASE INHIBITOR OF THE RICE, ORYZA SATIVA L. JAPONICA | | Descriptor: | ORYZACYSTATIN-I | | Authors: | Nagata, K, Kudo, N, Abe, K, Arai, S, Tanokura, M. | | Deposit date: | 2000-04-05 | | Release date: | 2001-01-10 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of oryzacystatin-I, a cysteine proteinase inhibitor of the rice, Oryza sativa L. japonica.
Biochemistry, 39, 2000
|
|
1F5V
 
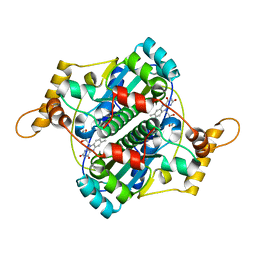 | | STRUCTURE AND SITE-DIRECTED MUTAGENESIS OF A FLAVOPROTEIN FROM ESCHERICHIA COLI THAT REDUCES NITROCOMPOUNDS. ALTERATION OF PYRIDINE NUCLEOTIDE BINDING BY A SINGLE AMINO ACID SUBSTITUTION | | Descriptor: | FLAVIN MONONUCLEOTIDE, OXYGEN-INSENSITIVE NADPH NITROREDUCTASE | | Authors: | Kobori, T, Sasaki, H, Lee, W.C, Zenno, S, Saigo, K, Murphy, M.E.P, Tanokura, M. | | Deposit date: | 2000-06-17 | | Release date: | 2001-02-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and site-directed mutagenesis of a flavoprotein from Escherichia coli that reduces nitrocompounds: alteration of pyridine nucleotide binding by a single amino acid substitution.
J.Biol.Chem., 276, 2001
|
|
3JRS
 
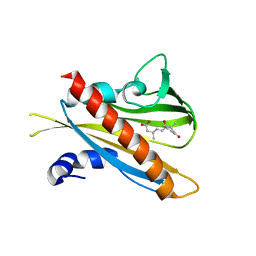 | | Crystal structure of (+)-ABA-bound PYL1 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Putative uncharacterized protein At5g46790 | | Authors: | Miyazono, K, Miyakawa, T, Sawano, Y, Kubota, K, Tanokura, M. | | Deposit date: | 2009-09-08 | | Release date: | 2009-11-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis of abscisic acid signalling
Nature, 462, 2009
|
|
3JRQ
 
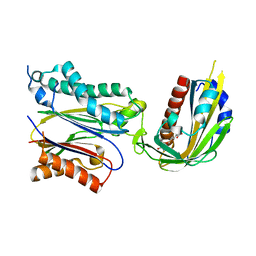 | | Crystal structure of (+)-ABA-bound PYL1 in complex with ABI1 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Protein phosphatase 2C 56, Putative uncharacterized protein At5g46790 | | Authors: | Miyazono, K, Miyakawa, T, Sawano, Y, Kubota, K, Tanokura, M. | | Deposit date: | 2009-09-08 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of abscisic acid signalling
Nature, 462, 2009
|
|
2RR7
 
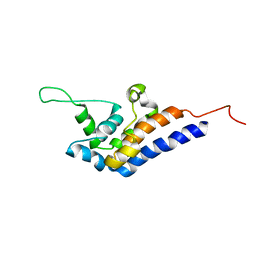 | | Microtubule Binding Domain of DYNEIN-C | | Descriptor: | Dynein heavy chain 9 | | Authors: | Kato, Y, Yagi, T, Ohki, S, Burgess, S, Honda, S, Kamiya, R, Tanokura, M. | | Deposit date: | 2010-06-04 | | Release date: | 2011-06-08 | | Last modified: | 2015-09-09 | | Method: | SOLUTION NMR | | Cite: | Structure of the microtubule-binding domain of flagellar dynein
Structure, 22, 2014
|
|
1IX5
 
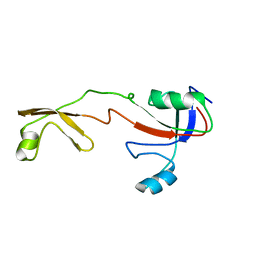 | | Solution structure of the Methanococcus thermolithotrophicus FKBP | | Descriptor: | FKBP | | Authors: | Suzuki, R, Nagata, K, Kawakami, M, Nemoto, N, Furutani, M, Adachi, K, Maruyama, T, Tanokura, M. | | Deposit date: | 2002-06-12 | | Release date: | 2003-06-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional Solution Structure of an Archaeal FKBP with a Dual Function of Peptidyl Prolyl cis-trans Isomerase and Chaperone-like Activities
J.MOL.BIOL., 328, 2003
|
|
5WUT
 
 | |
5GWT
 
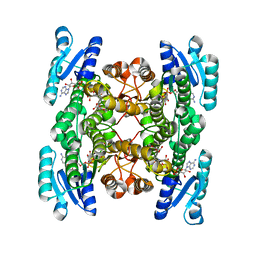 | | 4-hydroxyisoleucine dehydrogenase mutant complexed with NADH and succinate | | Descriptor: | 4-hydroxyisolecuine dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SUCCINIC ACID | | Authors: | Shi, X, Miyakawa, T, Nakamura, A, Tanokura, M. | | Deposit date: | 2016-09-13 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Engineering a short-chain dehydrogenase/reductase for the stereoselective production of (2S,3R,4S)-4-hydroxyisoleucine with three asymmetric centers.
Sci Rep, 7, 2017
|
|
5GWR
 
 | | 4-hydroxyisoleucine dehydrogenase complexed with NADH | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxyisolecuine dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Shi, X, Miyakawa, T, Nakamura, A, Tanokura, M. | | Deposit date: | 2016-09-13 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Engineering a short-chain dehydrogenase/reductase for the stereoselective production of (2S,3R,4S)-4-hydroxyisoleucine with three asymmetric centers
Sci Rep, 7, 2017
|
|
5GWS
 
 | | 4-hydroxyisoleucine dehydrogenase complexed with NADH and succinate | | Descriptor: | 4-hydroxyisolecuine dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SUCCINIC ACID | | Authors: | Shi, X, Miyakawa, T, Nakamura, A, Tanokura, M. | | Deposit date: | 2016-09-13 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Engineering a short-chain dehydrogenase/reductase for the stereoselective production of (2S,3R,4S)-4-hydroxyisoleucine with three asymmetric centers.
Sci Rep, 7, 2017
|
|
