3Q35
 
 | | Structure of the Rtt109-AcCoA/Vps75 complex and implications for chaperone-mediated histone acetylation | | Descriptor: | 1,2-ETHANEDIOL, ACETYL COENZYME *A, Histone acetyltransferase, ... | | Authors: | Tang, Y, Yuan, H, Meeth, K, Marmorstein, R. | | Deposit date: | 2010-12-21 | | Release date: | 2011-02-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the Rtt109-AcCoA/Vps75 Complex and Implications for Chaperone-Mediated Histone Acetylation.
Structure, 19, 2011
|
|
3Q33
 
 | | Structure of the Rtt109-AcCoA/Vps75 Complex and Implications for Chaperone-Mediated Histone Acetylation | | Descriptor: | 1,2-ETHANEDIOL, ACETYL COENZYME *A, HISTONE H3, ... | | Authors: | Tang, Y, Yuan, H, Meeth, K, Marmorstein, R. | | Deposit date: | 2010-12-21 | | Release date: | 2011-02-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Rtt109-AcCoA/Vps75 Complex and Implications for Chaperone-Mediated Histone Acetylation.
Structure, 19, 2011
|
|
2N42
 
 | | EC-NMR Structure of Human H-RasT35S mutant protein Determined by Combining Evolutionary Couplings (EC) and Sparse NMR Data | | Descriptor: | GTPase HRas | | Authors: | Tang, Y, Huang, Y.J, Hopf, T.A, Sander, C, Marks, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-16 | | Release date: | 2015-07-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination by combining sparse NMR data with evolutionary couplings.
Nat.Methods, 12, 2015
|
|
2N46
 
 | | EC-NMR Structure of Human H-RasT35S mutant protein Determined by Combining Evolutionary Couplings (EC) and Sparse NMR Data | | Descriptor: | GTPase HRas | | Authors: | Tang, Y, Huang, Y.J, Hopf, T.A, Sander, C, Marks, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-17 | | Release date: | 2015-07-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination by combining sparse NMR data with evolutionary couplings.
Nat.Methods, 12, 2015
|
|
2N44
 
 | | EC-NMR Structure of Escherichia coli Maltose-binding protein Determined by Combining Evolutionary Couplings (EC) and Sparse NMR Data. Northeast Structural Genomics Consortium target ER690 | | Descriptor: | Maltose-binding periplasmic protein | | Authors: | Tang, Y, Huang, Y.J, Hopf, T.A, Sander, C, Marks, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-16 | | Release date: | 2015-07-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination by combining sparse NMR data with evolutionary couplings.
Nat.Methods, 12, 2015
|
|
2N45
 
 | | EC-NMR Structure of Escherichia coli Maltose-binding protein Determined by Combining Evolutionary Couplings (EC) and Sparse NMR Data with a second set of RDC data simulated for an alternative alignment tensor. Northeast Structural Genomics Consortium target ER690 | | Descriptor: | Maltose-binding periplasmic protein | | Authors: | Tang, Y, Huang, Y.J, Hopf, T.A, Sander, C, Marks, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-17 | | Release date: | 2015-07-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination by combining sparse NMR data with evolutionary couplings.
Nat.Methods, 12, 2015
|
|
2N47
 
 | | EC-NMR Structure of Synechocystis sp. PCC 6803 Slr1183 Determined by Combining Evolutionary Couplings (EC) and Sparse NMR Data. Northeast Structural Genomics Consortium target SgR145 | | Descriptor: | Slr1183 protein | | Authors: | Tang, Y, Huang, Y.J, Hopf, T.A, Sander, C, Marks, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-17 | | Release date: | 2015-07-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination by combining sparse NMR data with evolutionary couplings.
Nat.Methods, 12, 2015
|
|
3DM7
 
 | |
1U5F
 
 | |
2N49
 
 | | EC-NMR Structure of Erwinia carotovora ECA1580 N-terminal Domain Determined by Combining Evolutionary Couplings (EC) and Sparse NMR Data. Northeast Structural Genomics Consortium target EwR156A | | Descriptor: | Putative cold-shock protein | | Authors: | Tang, Y, Huang, Y.J, Hopf, T.A, Sander, C, Marks, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-17 | | Release date: | 2015-07-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination by combining sparse NMR data with evolutionary couplings.
Nat.Methods, 12, 2015
|
|
2N4A
 
 | | EC-NMR Structure of Ralstonia metallidurans Rmet_5065 Determined by Combining Evolutionary Couplings (EC) and Sparse NMR Data. Northeast Structural Genomics Consortium target CrR115 | | Descriptor: | Uncharacterized protein | | Authors: | Tang, Y, Huang, Y.J, Hopf, T.A, Sander, C, Marks, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-17 | | Release date: | 2015-07-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination by combining sparse NMR data with evolutionary couplings.
Nat.Methods, 12, 2015
|
|
2N4B
 
 | | EC-NMR Structure of Ralstonia metallidurans Rmet_5065 Determined by Combining Evolutionary Couplings (EC) and Sparse NMR Data. Northeast Structural Genomics Consortium target CrR115 | | Descriptor: | Uncharacterized protein | | Authors: | Tang, Y, Huang, Y.J, Hopf, T.A, Sander, C, Marks, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-17 | | Release date: | 2015-07-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination by combining sparse NMR data with evolutionary couplings.
Nat.Methods, 12, 2015
|
|
2N4C
 
 | | EC-NMR Structure of Agrobacterium tumefaciens Atu1203 Determined by Combining Evolutionary Couplings (EC) and Sparse NMR Data. Northeast Structural Genomics Consortium target AtT10 | | Descriptor: | Uncharacterized protein Atu1203 | | Authors: | Tang, Y, Huang, Y.J, Hopf, T.A, Sander, C, Marks, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2015-06-17 | | Release date: | 2015-07-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination by combining sparse NMR data with evolutionary couplings.
Nat.Methods, 12, 2015
|
|
2N48
 
 | | EC-NMR Structure of Escherichia coli YiaD Determined by Combining Evolutionary Couplings (EC) and Sparse NMR Data. Northeast Structural Genomics Consortium target ER553 | | Descriptor: | Probable lipoprotein YiaD | | Authors: | Tang, Y, Huang, Y.J, Hopf, T.A, Sander, C, Marks, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-17 | | Release date: | 2015-07-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination by combining sparse NMR data with evolutionary couplings.
Nat.Methods, 12, 2015
|
|
2N4F
 
 | | EC-NMR Structure of Arabidopsis thaliana At2g32350 Determined by Combining Evolutionary Couplings (EC) and Sparse NMR Data. Northeast Structural Genomics Consortium target AR3433A | | Descriptor: | uncharacterized protein AR3433A | | Authors: | Tang, Y, Huang, Y.J, Hopf, T.A, Sander, C, Marks, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-17 | | Release date: | 2015-07-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination by combining sparse NMR data with evolutionary couplings.
Nat.Methods, 12, 2015
|
|
2N4D
 
 | | EC-NMR Structure of Agrobacterium tumefaciens Atu1203 Determined by Combining Evolutionary Couplings (EC) and Sparse NMR Data. Northeast Structural Genomics Consortium target AtT10 | | Descriptor: | Uncharacterized protein Atu1203 | | Authors: | Tang, Y, Huang, Y.J, Hopf, T.A, Sander, C, Marks, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-17 | | Release date: | 2015-07-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination by combining sparse NMR data with evolutionary couplings.
Nat.Methods, 12, 2015
|
|
3QM0
 
 | | Crystal structure of RTT109-AC-CoA complex | | Descriptor: | ACETYL COENZYME *A, Histone acetyltransferase RTT109, MERCURY (II) ION | | Authors: | Tang, Y, Marmorstein, R. | | Deposit date: | 2011-02-03 | | Release date: | 2011-02-16 | | Last modified: | 2017-08-02 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Fungal Rtt109 histone acetyltransferase is an unexpected structural homolog of metazoan p300/CBP.
Nat.Struct.Mol.Biol., 15, 2008
|
|
3R6G
 
 | |
3R7S
 
 | | Crystal Structure of Apo Caspase2 | | Descriptor: | Caspase-2 subunit p12, Caspase-2 subunit p18 | | Authors: | Tang, Y, Wells, J, Arkin, M. | | Deposit date: | 2011-03-22 | | Release date: | 2011-07-27 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | Structural and enzymatic insights into caspase-2 protein substrate recognition and catalysis.
J.Biol.Chem., 286, 2011
|
|
3R5J
 
 | |
3R6L
 
 | | Caspase-2 T380A bound with Ac-VDVAD-CHO | | Descriptor: | Caspase-2 subunit p12, Caspase-2 subunit p18, Peptide Inhibitor (ACE)VDVAD-CHO | | Authors: | Tang, Y, Wells, J, Arkin, M. | | Deposit date: | 2011-03-21 | | Release date: | 2011-07-27 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and enzymatic insights into caspase-2 protein substrate recognition and catalysis.
J.Biol.Chem., 286, 2011
|
|
3R7B
 
 | | Caspase-2 bound to one copy of Ac-DVAD-CHO | | Descriptor: | Caspase-2 subunit p12, Caspase-2 subunit p18, Peptide Inhibitor (ACE)DVAD-CHO | | Authors: | Tang, Y, Wells, J, Arkin, M. | | Deposit date: | 2011-03-22 | | Release date: | 2011-07-27 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and enzymatic insights into caspase-2 protein substrate recognition and catalysis.
J.Biol.Chem., 286, 2011
|
|
2L15
 
 | | Solution Structure of Cold Shock Protein CspA Using Combined NMR and CS-Rosetta method | | Descriptor: | Cold shock protein CspA | | Authors: | Tang, Y, Schneider, W.M, Shen, Y, Raman, S, Inouye, M, Baker, D, Roth, M.J, Montelione, G.T. | | Deposit date: | 2010-07-22 | | Release date: | 2010-09-15 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Fully automated high-quality NMR structure determination of small (2)H-enriched proteins.
J Struct Funct Genomics, 11, 2010
|
|
3R7N
 
 | | Caspase-2 bound with two copies of Ac-DVAD-CHO | | Descriptor: | Caspase-2 subunit p12, Caspase-2 subunit p18, Peptide Inhibitor (ACE)DVAD-CHO | | Authors: | Tang, Y, Wells, J, Arkin, M. | | Deposit date: | 2011-03-22 | | Release date: | 2011-07-27 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural and enzymatic insights into caspase-2 protein substrate recognition and catalysis.
J.Biol.Chem., 286, 2011
|
|
2LZK
 
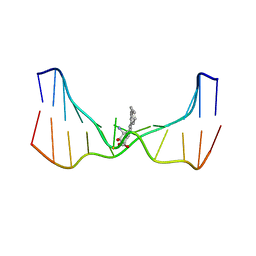 | | NMR solution structure of an N2-guanine DNA adduct derived from the potent tumorigen dibenzo[a,l]pyrene: Intercalation from the minor groove with ruptured Watson-Crick base pairing | | Descriptor: | (11S,12S,13S)-11,12,13,14-tetrahydronaphtho[1,2,3,4-pqr]tetraphene-11,12,13-triol, DNA (5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3') | | Authors: | Tang, Y, Liu, Z, Ding, S, Lin, C.H, Cai, Y, Rodriguez, F.A, Sayer, J.M, Jerina, D.M, Amin, S, Broyde, S, Geacintov, N.E. | | Deposit date: | 2012-10-04 | | Release date: | 2012-11-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Solution Structure of an N(2)-Guanine DNA Adduct Derived from the Potent Tumorigen Dibenzo[a,l]pyrene: Intercalation from the Minor Groove with Ruptured Watson-Crick Base Pairing.
Biochemistry, 51, 2012
|
|
