1GP8
 
 | | NMR SOLUTION STRUCTURE OF THE COAT PROTEIN-BINDING DOMAIN OF BACTERIOPHAGE P22 SCAFFOLDING PROTEIN | | Descriptor: | PROTEIN (SCAFFOLDING PROTEIN) | | Authors: | Sun, Y, Parker, M.H, Weigele, P, Casjens, S, Prevelige Jr, P.E, Krishna, N.R. | | Deposit date: | 1999-05-11 | | Release date: | 1999-05-17 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the coat protein-binding domain of the scaffolding protein from a double-stranded DNA virus.
J.Mol.Biol., 297, 2000
|
|
7R8E
 
 | | The structure of human ABCG1 E242Q complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHOLESTEROL, Isoform 4 of ATP-binding cassette sub-family G member 1, ... | | Authors: | Sun, Y, Li, X, Long, T. | | Deposit date: | 2021-06-26 | | Release date: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Molecular basis of cholesterol efflux via ABCG subfamily transporters.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7R8C
 
 | | The structure of human ABCG1 | | Descriptor: | Isoform 4 of ATP-binding cassette sub-family G member 1 | | Authors: | Sun, Y, Li, X, Long, T. | | Deposit date: | 2021-06-26 | | Release date: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular basis of cholesterol efflux via ABCG subfamily transporters.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7R87
 
 | | The structure of human ABCG5-WT/ABCG8-I419E | | Descriptor: | 2C7 Fab heavy chain, 2C7 Fab light chain, ATP-binding cassette sub-family G member 5, ... | | Authors: | Sun, Y, Li, X, Long, T. | | Deposit date: | 2021-06-26 | | Release date: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis of cholesterol efflux via ABCG subfamily transporters.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7R88
 
 | | The structure of human ABCG5-I529W/ABCG8-WT | | Descriptor: | 2C7 Fab heavy chain, 2C7 Fab light chain, ATP-binding cassette sub-family G member 5, ... | | Authors: | Sun, Y, Li, X, Long, T. | | Deposit date: | 2021-06-26 | | Release date: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis of cholesterol efflux via ABCG subfamily transporters.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7R8D
 
 | | The structure of human ABCG1 E242Q with cholesterol | | Descriptor: | CHOLESTEROL, Isoform 4 of ATP-binding cassette sub-family G member 1 | | Authors: | Sun, Y, Li, X, Long, T. | | Deposit date: | 2021-06-26 | | Release date: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular basis of cholesterol efflux via ABCG subfamily transporters.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7R89
 
 | | The structure of human ABCG5/ABCG8 purified from yeast | | Descriptor: | 2C7 Fab heavy chain, 2C7 Fab light chain, ATP-binding cassette sub-family G member 5, ... | | Authors: | Sun, Y, Li, X, Long, T. | | Deposit date: | 2021-06-26 | | Release date: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Molecular basis of cholesterol efflux via ABCG subfamily transporters.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7R8B
 
 | | The structure of human ABCG5/ABCG8 supplemented with cholesterol | | Descriptor: | 2C7 Fab heavy chain, 2C7 Fab light chain, ATP-binding cassette sub-family G member 5, ... | | Authors: | Sun, Y, Li, X, Long, T. | | Deposit date: | 2021-06-26 | | Release date: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis of cholesterol efflux via ABCG subfamily transporters.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7R8A
 
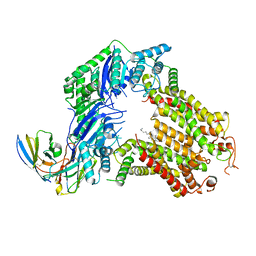 | | The structure of human ABCG5/ABCG8 purified from mammalian cells | | Descriptor: | 2C7 Fab heavy chain, 2C7 Fab light chain, ATP-binding cassette sub-family G member 5, ... | | Authors: | Sun, Y, Li, X, Long, T. | | Deposit date: | 2021-06-26 | | Release date: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular basis of cholesterol efflux via ABCG subfamily transporters.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6PPO
 
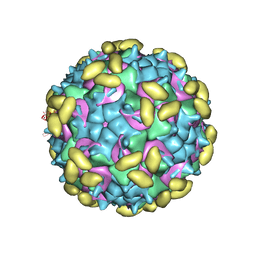 | | Rhinovirus C15 complexed with domain I of receptor CDHR3 | | Descriptor: | CALCIUM ION, Cadherin-related family member 3, Capsid protein VP1, ... | | Authors: | Sun, Y, Watters, K, Klose, T, Palmenberg, A.C. | | Deposit date: | 2019-07-08 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of rhinovirus C15a bound to its cadherin-related protein 3 receptor.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6PSF
 
 | | Rhinovirus C15 complexed with domains I and II of receptor CDHR3 | | Descriptor: | Cadherin-related family member 3, Capsid protein VP1, Capsid protein VP2, ... | | Authors: | Sun, Y, Watters, K, Klose, T, Palmenberg, A.C. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of rhinovirus C15a bound to its cadherin-related protein 3 receptor.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6V4X
 
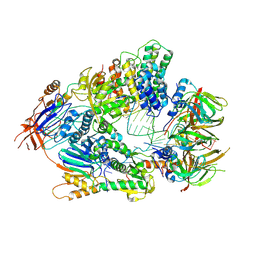 | | Cryo-EM structure of an active human histone pre-mRNA 3'-end processing machinery at 3.2 Angstrom resolution | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 2, Cleavage and polyadenylation specificity factor subunit 3, Small nuclear ribonucleoprotein E, ... | | Authors: | Sun, Y, Zhang, Y, Walz, T, Tong, L. | | Deposit date: | 2019-12-02 | | Release date: | 2020-02-19 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of an active human histone pre-mRNA 3'-end processing machinery.
Science, 367, 2020
|
|
2Q5Y
 
 | | Crystal Structure of the C-terminal domain of hNup98 | | Descriptor: | Nuclear pore complex protein Nup96, Nuclear pore complex protein Nup98 | | Authors: | Sun, Y, Guo, H.C. | | Deposit date: | 2007-06-03 | | Release date: | 2008-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural constraints on autoprocessing of the human nucleoporin Nup98.
Protein Sci., 17, 2008
|
|
2Q5X
 
 | |
3MGT
 
 | | Crystal structure of a H5-specific CTL epitope variant derived from H5N1 influenza virus in complex with HLA-A*0201 | | Descriptor: | 10-meric peptide from Hemagglutinin, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Sun, Y, Liu, J, Yang, M, Gao, F, Zhou, J, Kitamura, Y. | | Deposit date: | 2010-04-07 | | Release date: | 2010-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Identification and structural definition of H5-specific CTL epitopes restricted by HLA-A*0201 derived from the H5N1 subtype of influenza A viruses
J.Gen.Virol., 91, 2010
|
|
3MGO
 
 | | Crystal structure of a H5-specific CTL epitope derived from H5N1 influenza virus in complex with HLA-A*0201 | | Descriptor: | 10-meric peptide from Hemagglutinin, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Sun, Y, Liu, J, Yang, M, Gao, F, Zhou, J, Kitamura, Y. | | Deposit date: | 2010-04-07 | | Release date: | 2010-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Identification and structural definition of H5-specific CTL epitopes restricted by HLA-A*0201 derived from the H5N1 subtype of influenza A viruses
J.Gen.Virol., 91, 2010
|
|
6O5K
 
 | | Murine TRIM28 Bbox1 domain | | Descriptor: | Transcription intermediary factor 1-beta, ZINC ION | | Authors: | Sun, Y, Keown, J.R, Goldstone, D.C. | | Deposit date: | 2019-03-03 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Dissection of Oligomerization by the TRIM28 Tripartite Motif and the Interaction with Members of the Krab-ZFP Family.
J.Mol.Biol., 431, 2019
|
|
3IFU
 
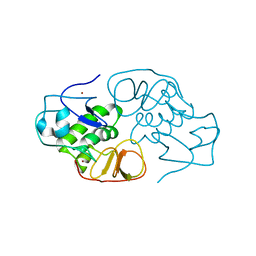 | | The Crystal Structure of Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) Leader Protease Nsp1 | | Descriptor: | Non-structural protein, ZINC ION | | Authors: | Sun, Y, Xue, F, Guo, Y, Ma, M, Lou, Z, Rao, Z. | | Deposit date: | 2009-07-26 | | Release date: | 2009-10-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Crystal structure of porcine reproductive and respiratory syndrome virus leader protease Nsp1alpha
J.Virol., 83, 2009
|
|
1LB8
 
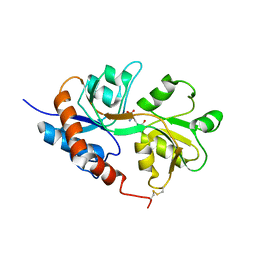 | | Crystal structure of the Non-desensitizing GluR2 ligand binding core mutant (S1S2J-L483Y) in complex with AMPA at 2.3 resolution | | Descriptor: | (S)-ALPHA-AMINO-3-HYDROXY-5-METHYL-4-ISOXAZOLEPROPIONIC ACID, Glutamate receptor 2 | | Authors: | Sun, Y, Olson, R, Horning, M, Armstrong, N, Mayer, M, Gouaux, E. | | Deposit date: | 2002-04-02 | | Release date: | 2002-06-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism of glutamate receptor desensitization.
Nature, 417, 2002
|
|
1LB9
 
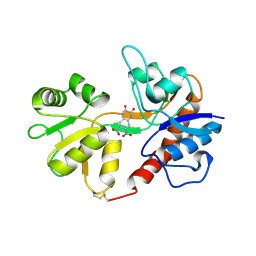 | | Crystal structure of the Non-desensitizing GluR2 ligand binding core mutant (S1S2J-L483Y) in complex with antagonist DNQX at 2.3 A resolution | | Descriptor: | 6,7-DINITROQUINOXALINE-2,3-DIONE, Glutamate receptor 2, SULFATE ION | | Authors: | Sun, Y, Olson, R, Horning, M, Armstrong, N, Mayer, M, Gouaux, E. | | Deposit date: | 2002-04-02 | | Release date: | 2002-06-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism of glutamate receptor desensitization.
Nature, 417, 2002
|
|
1LBB
 
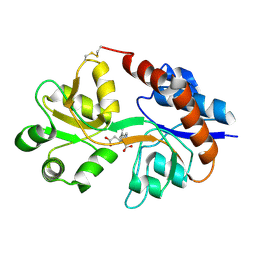 | | Crystal structure of the GluR2 ligand binding domain mutant (S1S2J-N754D) in complex with kainate at 2.1 A resolution | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamine receptor 2 | | Authors: | Sun, Y, Olson, R, Horning, M, Armstrong, N, Mayer, M, Gouaux, E. | | Deposit date: | 2002-04-02 | | Release date: | 2002-06-05 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of glutamate receptor desensitization.
Nature, 417, 2002
|
|
1LBC
 
 | | Crystal structure of GluR2 ligand binding core (S1S2J-N775S) in complex with cyclothiazide (CTZ) as well as glutamate at 1.8 A resolution | | Descriptor: | CYCLOTHIAZIDE, GLUTAMIC ACID, Glutamine Receptor 2, ... | | Authors: | Sun, Y, Olson, R, Horning, M, Armstrong, N, Mayer, M, Gouaux, E. | | Deposit date: | 2002-04-02 | | Release date: | 2002-05-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of glutamate receptor desensitization.
Nature, 417, 2002
|
|
4I42
 
 | | E.coli. 1,4-dihydroxy-2-naphthoyl coenzyme A synthase (ecMenB) in complex with 1-hydroxy-2-naphthoyl-CoA | | Descriptor: | 1,2-ETHANEDIOL, 1,4-Dihydroxy-2-naphthoyl-CoA synthase, 1-hydroxy-2-naphthoyl-CoA, ... | | Authors: | Sun, Y, Song, H, Li, J, Li, Y, Jiang, M, Zhou, J, Guo, Z. | | Deposit date: | 2012-11-27 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Structural basis of the induced-fit mechanism of 1,4-dihydroxy-2-naphthoyl coenzyme A synthase from the crotonase fold superfamily
Plos One, 8, 2013
|
|
2GP8
 
 | | NMR SOLUTION STRUCTURE OF THE COAT PROTEIN-BINDING DOMAIN OF BACTERIOPHAGE P22 SCAFFOLDING PROTEIN | | Descriptor: | PROTEIN (SCAFFOLDING PROTEIN) | | Authors: | Sun, Y, Parker, M.H, Weigele, P, Casjens, S, Prevelige Jr, P.E, Krishna, N.R. | | Deposit date: | 1999-05-11 | | Release date: | 1999-05-17 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the coat protein-binding domain of the scaffolding protein from a double-stranded DNA virus.
J.Mol.Biol., 297, 2000
|
|
2N1I
 
 | |
