5KTZ
 
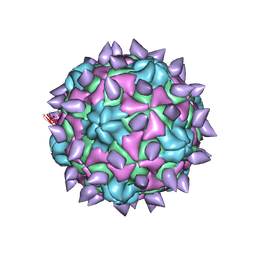 | | expanded poliovirus in complex with VHH 12B | | Descriptor: | Genome polyprotein, VHH 12B, VP1, ... | | Authors: | Strauss, M, Schotte, L, Filman, D.J, Hogle, J.M. | | Deposit date: | 2016-07-12 | | Release date: | 2016-11-02 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-electron Microscopy Structures of Expanded Poliovirus with VHHs Sample the Conformational Repertoire of the Expanded State.
J. Virol., 91, 2017
|
|
5KU0
 
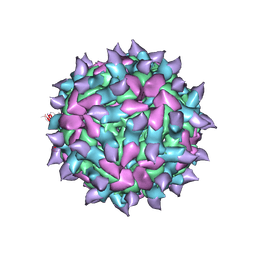 | | expanded poliovirus in complex with VHH 17B | | Descriptor: | VHH 17B, VP1, VP2, ... | | Authors: | Strauss, M, Schotte, L, Filman, D.J, Hogle, J.M. | | Deposit date: | 2016-07-12 | | Release date: | 2016-11-02 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-electron Microscopy Structures of Expanded Poliovirus with VHHs Sample the Conformational Repertoire of the Expanded State.
J. Virol., 91, 2017
|
|
5KU2
 
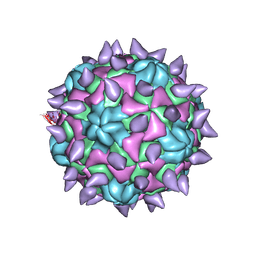 | | expanded poliovirus in complex with VHH 7A | | Descriptor: | VHH 7A, VP1, VP2, ... | | Authors: | Strauss, M, Schotte, L, Filman, D.J, Hogle, J.M. | | Deposit date: | 2016-07-12 | | Release date: | 2016-11-02 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-electron Microscopy Structures of Expanded Poliovirus with VHHs Sample the Conformational Repertoire of the Expanded State.
J. Virol., 91, 2017
|
|
5KWL
 
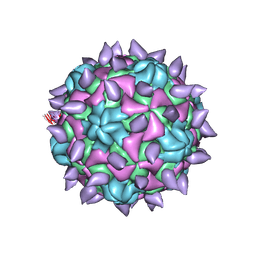 | | expanded poliovirus in complex with VHH 10E | | Descriptor: | VHH 10E, VP1, VP2, ... | | Authors: | Strauss, M, Schotte, L, Filman, D.J, Hogle, J.M. | | Deposit date: | 2016-07-18 | | Release date: | 2016-11-02 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-electron Microscopy Structures of Expanded Poliovirus with VHHs Sample the Conformational Repertoire of the Expanded State.
J. Virol., 91, 2017
|
|
2MI6
 
 | |
3JBE
 
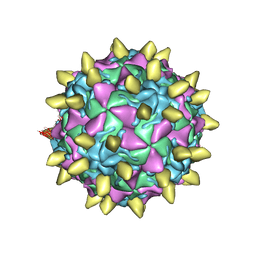 | | Complex of poliovirus with VHH PVSS8A | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Strauss, M, Schotte, L, Thys, B, Filman, D.J, Hogle, J.M. | | Deposit date: | 2015-08-26 | | Release date: | 2016-01-27 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Five of Five VHHs Neutralizing Poliovirus Bind the Receptor-Binding Site.
J.Virol., 90, 2016
|
|
3J8F
 
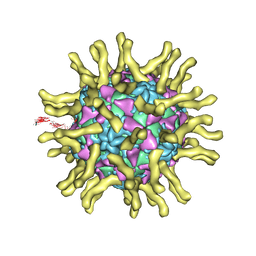 | | Cryo-EM reconstruction of poliovirus-receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid protein VP1, ... | | Authors: | Strauss, M, Filman, D.J, Belnap, D.M, Cheng, N, Noel, R.T, Hogle, J.M. | | Deposit date: | 2014-10-20 | | Release date: | 2015-02-11 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Nectin-Like Interactions between Poliovirus and Its Receptor Trigger Conformational Changes Associated with Cell Entry.
J.Virol., 89, 2015
|
|
3JBF
 
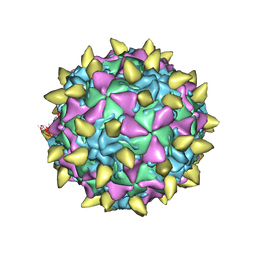 | | Complex of poliovirus with VHH PVSP19B | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Strauss, M, Schotte, L, Thys, B, Filman, D.J, Hogle, J.M. | | Deposit date: | 2015-08-26 | | Release date: | 2016-01-27 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Five of Five VHHs Neutralizing Poliovirus Bind the Receptor-Binding Site.
J.Virol., 90, 2016
|
|
3JBD
 
 | | Complex of poliovirus with VHH PVSP6A | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Strauss, M, Schotte, L, Thys, B, Filman, D.J, Hogle, J.M. | | Deposit date: | 2015-08-26 | | Release date: | 2016-01-27 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Five of Five VHHs Neutralizing Poliovirus Bind the Receptor-Binding Site.
J.Virol., 90, 2016
|
|
3JBC
 
 | | Complex of Poliovirus with VHH PVSP29F | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Strauss, M, Schotte, L, Thys, B, Filman, D.J, Hogle, J.M. | | Deposit date: | 2015-08-26 | | Release date: | 2016-01-27 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Five of Five VHHs Neutralizing Poliovirus Bind the Receptor-Binding Site.
J.Virol., 90, 2016
|
|
3JBG
 
 | | Complex of poliovirus with VHH PVSS21E | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Strauss, M, Schotte, L, Thys, B, Filman, D.J, Hogle, J.M. | | Deposit date: | 2015-08-26 | | Release date: | 2016-01-27 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Five of Five VHHs Neutralizing Poliovirus Bind the Receptor-Binding Site.
J.Virol., 90, 2016
|
|
3J9F
 
 | | Poliovirus complexed with soluble, deglycosylated poliovirus receptor (Pvr) at 4 degrees C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITIC ACID, ... | | Authors: | Strauss, M, Filman, D.J, Belnap, D.M, Cheng, N, Noel, R.T, Hogle, J.M. | | Deposit date: | 2015-01-15 | | Release date: | 2015-02-11 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Nectin-Like Interactions between Poliovirus and Its Receptor Trigger Conformational Changes Associated with Cell Entry.
J.Virol., 89, 2015
|
|
7THX
 
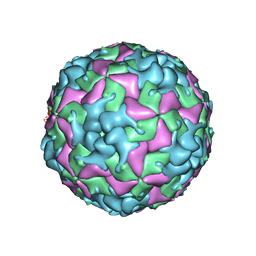 | | Cryo-EM structure of W6 possum enterovirus | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Wang, I, Jayawardena, N, Strauss, M, Bostina, M. | | Deposit date: | 2022-01-12 | | Release date: | 2022-03-09 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Cryo-EM Structure of a Possum Enterovirus.
Viruses, 14, 2022
|
|
7QID
 
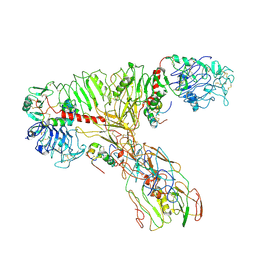 | | tentative model of the human insulin receptor ectodomain bound by three insulin | | Descriptor: | Insulin, Insulin receptor, Isoform Short of Insulin receptor | | Authors: | Gutman, T, Schaefer, I.B, Poojari, C.S, Vattulainen, I, Strauss, M, Coskun, U. | | Deposit date: | 2021-12-14 | | Release date: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Cryo-EM structure of the complete and ligand-saturated insulin receptor ectodomain
J Cell Biol, 219, 2020
|
|
8FXH
 
 | |
8FXI
 
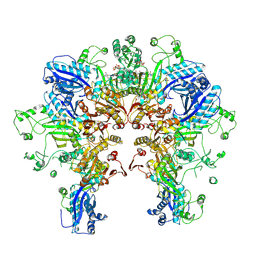 | | Cryo-EM structure of Stanieria sp. CphA2 in complex with ADPCP and 4x(beta-Asp-Arg) | | Descriptor: | 1-[(4-aminopyrimidin-5-yl)amino]-2,5-anhydro-1-deoxy-6-O-[(S)-hydroxy{[(R)-hydroxy(phosphonomethyl)phosphoryl]oxy}phosphoryl]-D-allitol, 4x(beta-Asp-Arg), MAGNESIUM ION, ... | | Authors: | Markus, L.M, Sharon, I, Strauss, M, Schmeing, T.M. | | Deposit date: | 2023-01-24 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure and function of a hexameric cyanophycin synthetase 2.
Protein Sci., 32, 2023
|
|
7AT8
 
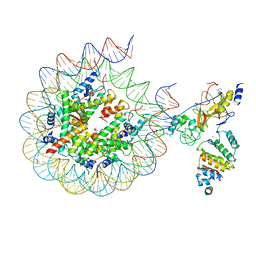 | | Histone H3 recognition by nucleosome-bound PRC2 subunit EZH2. | | Descriptor: | Histone H2A, Histone H2B 1.1, Histone H3.2, ... | | Authors: | Finogenova, K, Benda, C, Schaefer, I.B, Poepsel, S, Strauss, M, Mueller, J. | | Deposit date: | 2020-10-29 | | Release date: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural basis for PRC2 decoding of active histone methylation marks H3K36me2/3.
Elife, 9, 2020
|
|
7O3X
 
 | | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Protein sll0617 | | Authors: | Gupta, T.K, Klumpe, S, Gries, K, Strauss, M, Rudack, T, Schuller, J.M, Schroda, M, Engel, B.D. | | Deposit date: | 2021-04-03 | | Release date: | 2021-06-30 | | Last modified: | 2021-07-21 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity.
Cell, 184, 2021
|
|
7O40
 
 | | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Protein sll0617 | | Authors: | Gupta, T.K, Klumpe, S, Gries, K, Strauss, M, Rudack, T, Schuller, J.M, Schroda, M, Engel, B.D. | | Deposit date: | 2021-04-03 | | Release date: | 2021-06-30 | | Last modified: | 2021-07-21 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity.
Cell, 184, 2021
|
|
7O3Y
 
 | | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Protein sll0617 | | Authors: | Gupta, T.K, Klumpe, S, Gries, K, Strauss, M, Rudack, T, Schuller, J.M, Schroda, M, Engel, B.D. | | Deposit date: | 2021-04-03 | | Release date: | 2021-06-30 | | Last modified: | 2021-07-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity.
Cell, 184, 2021
|
|
7O3Z
 
 | | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Protein sll0617 | | Authors: | Gupta, T.K, Klumpe, S, Gries, K, Strauss, M, Rudack, T, Schuller, J.M, Schroda, M, Engel, B.D. | | Deposit date: | 2021-04-03 | | Release date: | 2021-06-30 | | Last modified: | 2021-07-21 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity.
Cell, 184, 2021
|
|
7O3W
 
 | | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Protein sll0617 | | Authors: | Gupta, T.K, Klumpe, S, Gries, K, Strauss, M, Rudack, T, Schuller, J.M, Schroda, M, Engel, B.D. | | Deposit date: | 2021-04-03 | | Release date: | 2021-06-30 | | Last modified: | 2021-07-21 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity.
Cell, 184, 2021
|
|
6NEF
 
 | | Outer Membrane Cytochrome S Filament from Geobacter Sulfurreducens | | Descriptor: | C-type cytochrome OmcS, HEME C, MAGNESIUM ION | | Authors: | Filman, D.J, Marino, S.F, Ward, J.E, Yang, L, Mester, Z, Bullitt, E, Lovley, D.R, Strauss, M. | | Deposit date: | 2018-12-17 | | Release date: | 2019-07-03 | | Last modified: | 2019-09-11 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Cryo-EM reveals the structural basis of long-range electron transport in a cytochrome-based bacterial nanowire.
Commun Biol, 2, 2019
|
|
6SOF
 
 | | human insulin receptor ectodomain bound by 4 insulin | | Descriptor: | Insulin, Insulin receptor | | Authors: | Gutmann, T, Schaefer, I.B, Poojari, C.S, Vattulainen, I, Strauss, M, Coskun, U. | | Deposit date: | 2019-08-29 | | Release date: | 2019-11-13 | | Last modified: | 2020-05-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structure of the complete and ligand-saturated insulin receptor ectodomain.
J.Cell Biol., 219, 2020
|
|
8CXP
 
 | | Characterisation of a Seneca Valley Virus Thermostable Mutant | | Descriptor: | Capsid protein VP1, Capsid protein VP3, VP2, ... | | Authors: | Jayawardena, N, Bostina, M, Strauss, M. | | Deposit date: | 2022-05-22 | | Release date: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | Characterisation of a Seneca Valley virus thermostable mutant.
Virology, 575, 2022
|
|
