1HJD
 
 | |
1HJ0
 
 | | Thymosin beta9 | | Descriptor: | THYMOSIN BETA9 | | Authors: | Stoll, R, Voelter, W, Holak, T.A. | | Deposit date: | 2001-01-05 | | Release date: | 2002-01-04 | | Last modified: | 2020-01-15 | | Method: | SOLUTION NMR | | Cite: | Conformation of Thymosin Beta9 in Water/Fluoroalcohol Solution Determined by NMR Spectroscopy
Biopolymers, 41, 1997
|
|
2PRT
 
 | | Structure of the Wilms Tumor Suppressor Protein Zinc Finger Domain Bound to DNA | | Descriptor: | DNA (5'-D(*CP*AP*GP*AP*CP*GP*CP*CP*CP*CP*CP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*GP*GP*GP*GP*GP*CP*GP*TP*CP*TP*G)-3'), Wilms tumor 1, ... | | Authors: | Stoll, R, Lee, B.M, Debler, E.W, Laity, J.H, Wilson, I.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2007-05-04 | | Release date: | 2008-03-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structure of the Wilms tumor suppressor protein zinc finger domain bound to DNA
J.Mol.Biol., 372, 2007
|
|
2JP9
 
 | | Structure of the Wilms Tumor Suppressor Protein Zinc Finger Domain Bound to DNA | | Descriptor: | DNA (5'-D(P*DCP*DGP*DCP*DGP*DGP*DGP*DGP*DGP*DCP*DGP*DTP*DCP*DTP*DGP*DCP*DGP*DC)-3'), DNA (5'-D(P*DGP*DCP*DGP*DCP*DAP*DGP*DAP*DCP*DGP*DCP*DCP*DCP*DCP*DCP*DGP*DCP*DG)-3'), Wilms tumor 1, ... | | Authors: | Stoll, R, Lee, B.M, Debler, E.W, Laity, J.H, Wilson, I.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2007-04-30 | | Release date: | 2007-10-30 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of the wilms tumor suppressor protein zinc finger domain bound to DNA
J.Mol.Biol., 372, 2007
|
|
2JPA
 
 | | Structure of the Wilms Tumor Suppressor Protein Zinc Finger Domain Bound to DNA | | Descriptor: | DNA (5'-D(P*DCP*DAP*DGP*DAP*DCP*DGP*DCP*DCP*DCP*DCP*DCP*DGP*DCP*DG)-3'), DNA (5'-D(P*DCP*DGP*DCP*DGP*DGP*DGP*DGP*DGP*DCP*DGP*DTP*DCP*DTP*DG)-3'), Wilms tumor 1, ... | | Authors: | Stoll, R, Lee, B.M, Debler, E.W, Laity, J.H, Wilson, I.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2007-05-01 | | Release date: | 2007-10-30 | | Last modified: | 2020-02-05 | | Method: | SOLUTION NMR | | Cite: | Structure of the wilms tumor suppressor protein zinc finger domain bound to DNA
J.Mol.Biol., 372, 2007
|
|
2L0X
 
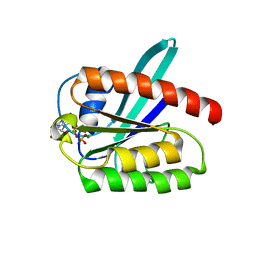 | | Solution structure of the 21 kDa GTPase RHEB bound to GDP | | Descriptor: | GTP-binding protein Rheb, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Stoll, R, Heumann, R, Berghaus, C, Kock, G. | | Deposit date: | 2010-07-19 | | Release date: | 2010-08-04 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Ras homolog enriched in brain (Rheb) enhances apoptotic signaling.
J.Biol.Chem., 285, 2010
|
|
7R3M
 
 | |
5IXB
 
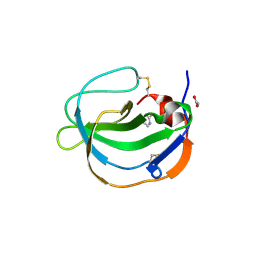 | | Structure of human Melanoma Inhibitory Activity (MIA) Protein in complex with Pyrimidin-2-amine | | Descriptor: | ACETATE ION, Melanoma-derived growth regulatory protein, PYRIMIDIN-2-AMINE | | Authors: | Yip, K.T, Gasper, R, Zhong, X.Y, Seibel, N, Puetz, S, Autzen, J, Scherkenbeck, J, Hofmann, E, Stoll, R. | | Deposit date: | 2016-03-23 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Small Molecules Antagonise the MIA-Fibronectin Interaction in Malignant Melanoma.
Sci Rep, 6, 2016
|
|
8RHR
 
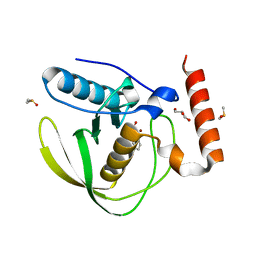 | | E.coli Peptide Deformylase with bound inhibitor BB4 | | Descriptor: | 2-(5-bromo-1H-indol-3-yl)-N-hydroxyacetamide, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Kirschner, H, Stoll, R, Hofmann, E. | | Deposit date: | 2023-12-16 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Toward More Selective Antibiotic Inhibitors: A Structural View of the Complexed Binding Pocket of E. coli Peptide Deformylase.
J.Med.Chem., 2024
|
|
6HNF
 
 | | Structure in solution of human fibronectin type III-domain 14 | | Descriptor: | Fibronectin | | Authors: | Zhong, X, Arnolds, O, Krenczyk, O, Gajewski, J, Puetz, S, Herrmann, C, Stoll, R. | | Deposit date: | 2018-09-14 | | Release date: | 2018-10-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Structure in Solution of Fibronectin Type III Domain 14 Reveals Its Synergistic Heparin Binding Site.
Biochemistry, 57, 2018
|
|
8OFE
 
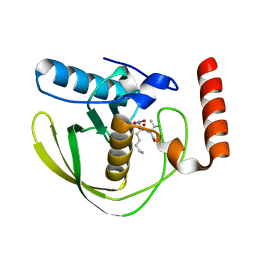 | | E.coli Peptide Deformylase with bound inhibitor | | Descriptor: | (2R)-2-[[methanoyl(oxidanyl)amino]methyl]-N-[(2S)-3-methyl-1-oxidanylidene-1-[2-(sulfanylmethyl)pyrrolidin-1-yl]butan-2-yl]heptanamide, Peptide deformylase, ZINC ION | | Authors: | Kirschner, H, Stoll, R, Hofmann, E. | | Deposit date: | 2023-03-15 | | Release date: | 2023-12-27 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Insights into Antibacterial Payload Release from Gold Nanoparticles Bound to E. coli Peptide Deformylase.
Chemmedchem, 19, 2024
|
|
4QNQ
 
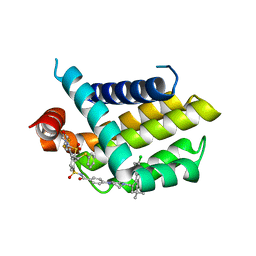 | |
7NHQ
 
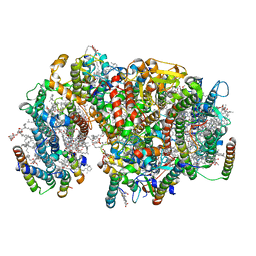 | | Structure of PSII-I prime (PSII with Psb28, and Psb34) | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, ... | | Authors: | Zabret, J, Bohn, S, Schuller, S.K, Arnolds, O, Chan, A, Tajkhorshid, E, Stoll, R, Engel, B.D, Rudack, T, Schuller, J.M, Nowaczyk, M.M. | | Deposit date: | 2021-02-11 | | Release date: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structural insights into photosystem II assembly.
Nat.Plants, 7, 2021
|
|
7NHP
 
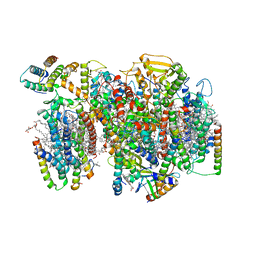 | | Structure of PSII-I (PSII with Psb27, Psb28, and Psb34) | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, ... | | Authors: | Zabret, J, Bohn, S, Schuller, S.K, Arnolds, O, Chan, A, Tajkhorshid, E, Stoll, R, Engel, B.D, Rudack, T, Schuller, J.M, Nowaczyk, M.M. | | Deposit date: | 2021-02-11 | | Release date: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Structural insights into photosystem II assembly.
Nat.Plants, 7, 2021
|
|
7NHO
 
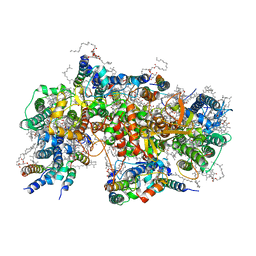 | | Structure of PSII-M | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, ... | | Authors: | Zabret, J, Bohn, S, Schuller, S.K, Arnolds, O, Chan, A, Tajkhorshid, E, Stoll, R, Engel, B.D, Rudack, T, Schuller, J.M, Nowaczyk, M.M. | | Deposit date: | 2021-02-11 | | Release date: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural insights into photosystem II assembly.
Nat.Plants, 7, 2021
|
|
6GBE
 
 | |
6GBD
 
 | |
2N5U
 
 | |
2K0C
 
 | | Zinc-finger 2 of Nup153 | | Descriptor: | Nuclear pore complex protein Nup153, ZINC ION | | Authors: | Bangert, J.A, Schrader, N, Vetter, I.R, Stoll, R. | | Deposit date: | 2008-01-31 | | Release date: | 2008-07-01 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | The Crystal Structure of the Ran-Nup153ZnF2 Complex: a General Ran Docking Site at the Nuclear Pore Complex.
Structure, 16, 2008
|
|
2KND
 
 | | Psb27 structure from Synechocystis | | Descriptor: | Photosystem II 11 kDa protein | | Authors: | Cormann, K.U, Nowaczyk, M.M, Bangert, J.-A, Ikeuchi, M, Roegner, M, Stoll, R. | | Deposit date: | 2009-08-21 | | Release date: | 2009-09-01 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure of Psb27 in solution: implications for transient binding to photosystem II during biogenesis and repair
Biochemistry, 48, 2009
|
|
2MXA
 
 | | Solution structure of the NDH-1 complex subunit CupS from Thermosynechococcus elongatus | | Descriptor: | NDH-1 complex sensory subunit CupS | | Authors: | Korste, A, Wulfhorst, H, Ikegami, T, Nowaczyk, M.M, Stoll, R. | | Deposit date: | 2014-12-17 | | Release date: | 2015-06-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the NDH-1 complex subunit CupS from Thermosynechococcus elongatus.
Biochim.Biophys.Acta, 1847, 2015
|
|
6X8O
 
 | | BimBH3 peptide tetramer | | Descriptor: | Bcl-2-like protein 11, THIOCYANATE ION | | Authors: | Robin, A.Y, Westphal, D, Uson, I, Czabotar, P.E. | | Deposit date: | 2020-06-01 | | Release date: | 2020-09-09 | | Last modified: | 2021-02-17 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Biophysical Characterization of Pro-apoptotic BimBH3 Peptides Reveals an Unexpected Capacity for Self-Association.
Structure, 29, 2021
|
|
3CH5
 
 | | The crystal structure of the RanGDP-Nup153ZnF2 complex | | Descriptor: | Fragment of Nuclear pore complex protein Nup153, GTP-binding nuclear protein Ran, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Vetter, I.R, Schrader, N. | | Deposit date: | 2008-03-07 | | Release date: | 2008-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of the Ran-Nup153ZnF2 Complex: a General Ran Docking Site at the Nuclear Pore Complex
Structure, 16, 2008
|
|
