1A3Y
 
 | |
6ZIG
 
 | |
6ZIH
 
 | |
6ZJJ
 
 | |
4RGG
 
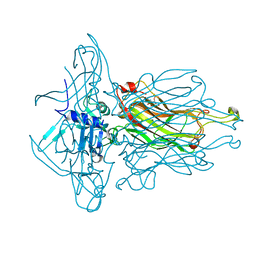 | |
2BSE
 
 | | Structure of Lactococcal Bacteriophage p2 Receptor Binding Protein in complex with a llama VHH domain | | Descriptor: | LLAMA IMMUNOGLOBULIN, RECEPTOR BINDING PROTEIN | | Authors: | Spinelli, S, Desmyter, A, Verrips, C.T, de Haard, H.J.W, Moineau, S, Cambillau, C. | | Deposit date: | 2005-05-20 | | Release date: | 2005-11-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Lactococcal Bacteriophage P2 Receptor Binding Protein Structure Suggests a Common Ancestor Gene with Bacterial and Mammalian Viruses.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2BSD
 
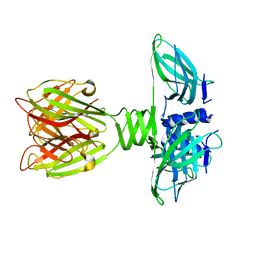 | | Structure of Lactococcal Bacteriophage p2 Receptor Binding Protein | | Descriptor: | RECEPTOR BINDING PROTEIN | | Authors: | Spinelli, S, Desmyter, A, Verrips, C.T, Dehaard, H.J.W, Moineau, S, Cambillau, C. | | Deposit date: | 2005-05-20 | | Release date: | 2005-11-02 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Lactococcal Bacteriophage P2 Receptor-Binding Protein Structure Suggests a Common Ancestor Gene with Bacterial and Mammalian Viruses.
Nat.Struct.Mol.Biol., 13, 2006
|
|
3R72
 
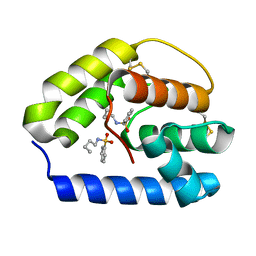 | | Apis mellifera odorant binding protein 5 | | Descriptor: | N-BUTYL-BENZENESULFONAMIDE, Odorant binding protein ASP5 | | Authors: | Spinelli, S, Iovinella, I, Lagarde, A, Tegoni, M, Pelosi, P, Cambillau, C. | | Deposit date: | 2011-03-22 | | Release date: | 2012-04-04 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structure of Apis mellifera odorant binding protein 5
To be Published
|
|
1HCV
 
 | |
1SJV
 
 | | Three-Dimensional Structure of a Llama VHH Domain Swapping | | Descriptor: | r9 | | Authors: | Spinelli, S, Desmyter, A, Frenken, L, Verrips, T, Cambillau, C. | | Deposit date: | 2004-03-04 | | Release date: | 2004-06-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Domain swapping of a llama VHH domain builds a crystal-wide beta-sheet structure.
Febs Lett., 564, 2004
|
|
1QD0
 
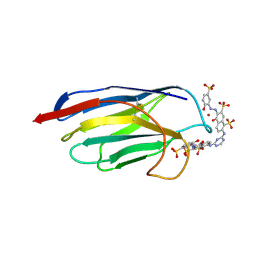 | | CAMELID HEAVY CHAIN VARIABLE DOMAINS PROVIDE EFFICIENT COMBINING SITES TO HAPTENS | | Descriptor: | 3-HYDROXY-7-(4-{1-[2-HYDROXY-3-(2-HYDROXY-5-SULFO-PHENYLAZO)-BENZYL]-2-SULFO-ETHYLAMINO}-[1,2,5]TRIAZIN-2-YLAMINO)-2-(2-HYDROXY-5-SULFO-PHENYLAZO)-NAPTHALENE-1,8-DISULFONIC ACID, COPPER (II) ION, VHH-R2 ANTI-RR6 ANTIBODY | | Authors: | Spinelli, S, Frenken, L.G.J, Hermans, P, Verrips, T, Brown, K, Tegoni, M, Cambillau, C. | | Deposit date: | 1999-07-08 | | Release date: | 2000-07-19 | | Last modified: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Camelid heavy-chain variable domains provide efficient combining sites to haptens.
Biochemistry, 39, 2000
|
|
1G1K
 
 | | COHESIN MODULE FROM THE CELLULOSOME OF CLOSTRIDIUM CELLULOLYTICUM | | Descriptor: | SCAFFOLDING PROTEIN | | Authors: | Spinelli, S, Fierobe, H.-P, Belaich, A, Belaich, J.-P, Henrissat, B, Cambillau, C. | | Deposit date: | 2000-10-12 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a cohesin module from Clostridium cellulolyticum: implications for dockerin recognition.
J.Mol.Biol., 304, 2000
|
|
1GM6
 
 | | 3-D STRUCTURE OF A SALIVARY LIPOCALIN FROM BOAR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, GLYCEROL, ... | | Authors: | Spinelli, S, Vincent, F, Pelosi, P, Tegoni, M, Cambillau, C. | | Deposit date: | 2001-09-11 | | Release date: | 2002-05-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Boar Salivary Lipocalin. Three-Dimensional X-Ray Structure and Androsterol/Androstenone Docking Simulations.
Eur.J.Biochem., 269, 2002
|
|
1HHP
 
 | |
3S0D
 
 | | Apis mellifera OBP 14 in complex with the citrus odorant citralva (3,7-dimethylocta-2,6-dienenitrile) | | Descriptor: | (2Z)-3,7-dimethylocta-2,6-dienenitrile, OBP14 | | Authors: | Spinelli, S, Lagarde, A, Iovinella, I, Tegoni, M, Pelosi, P, Cambillau, C. | | Deposit date: | 2011-05-13 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Crystal structure of Apis mellifera OBP14, a C-minus odorant-binding protein, and its complexes with odorant molecules.
Insect Biochem.Mol.Biol., 42, 2012
|
|
3S0E
 
 | | Apis mellifera OBP14 in complex with the odorant eugenol (2-methoxy-4(2-propenyl)-phenol) | | Descriptor: | 2-methoxy-4-(prop-2-en-1-yl)phenol, OBP14 | | Authors: | Spinelli, S, Lagarde, A, Iovinella, I, Tegoni, M, Pelosi, P, Cambillau, C. | | Deposit date: | 2011-05-13 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Apis mellifera OBP14, a C-minus odorant-binding protein, and its complexes with odorant molecules.
Insect Biochem.Mol.Biol., 42, 2012
|
|
3S0B
 
 | | Apis mellifera OBP14 in complex with the fluorescent probe 1-N-phenylnaphthylamine (NPN) | | Descriptor: | N-phenylnaphthalen-1-amine, OBP14 | | Authors: | Spinelli, S, Lagarde, A, Iovinella, I, Tegoni, M, Pelosi, P, Cambillau, C. | | Deposit date: | 2011-05-13 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Crystal structure of Apis mellifera OBP14, a C-minus odorant-binding protein, and its complexes with odorant molecules.
Insect Biochem.Mol.Biol., 42, 2012
|
|
3S0F
 
 | | Apis mellifera OBP14 native apo, crystal form 2 | | Descriptor: | OBP14 | | Authors: | Spinelli, S, Lagarde, A, Iovinella, I, Tegoni, M, Pelosi, P, Cambillau, C. | | Deposit date: | 2011-05-13 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of Apis mellifera OBP14, a C-minus odorant-binding protein, and its complexes with odorant molecules.
Insect Biochem.Mol.Biol., 42, 2012
|
|
3S0A
 
 | | Apis mellifera OBP14, native apo-protein | | Descriptor: | OBP14 | | Authors: | Spinelli, S, Lagarde, A, Iovinella, I, Tegoni, M, Pelosi, P, Cambillau, C. | | Deposit date: | 2011-05-13 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structure of Apis mellifera OBP14, a C-minus odorant-binding protein, and its complexes with odorant molecules.
Insect Biochem.Mol.Biol., 42, 2012
|
|
3S0G
 
 | | Apis mellifera OBP 14 double mutant Gln44Cys, His97Cys | | Descriptor: | OBP14 | | Authors: | Spinelli, S, Lagarde, A, Iovinella, I, Tegoni, M, Pelosi, P, Cambillau, C. | | Deposit date: | 2011-05-13 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Apis mellifera OBP14, a C-minus odorant-binding protein, and its complexes with odorant molecules.
Insect Biochem.Mol.Biol., 42, 2012
|
|
3RZS
 
 | | Apis mellifera OBP14 in complex with Ta6Br14 | | Descriptor: | HEXATANTALUM DODECABROMIDE, OBP14 | | Authors: | Spinelli, S, Lagarde, A, Iovinella, I, Tegoni, M, Pelosi, P, Cambillau, C. | | Deposit date: | 2011-05-12 | | Release date: | 2011-11-30 | | Last modified: | 2012-01-11 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of Apis mellifera OBP14, a C-minus odorant-binding protein, and its complexes with odorant molecules.
Insect Biochem.Mol.Biol., 42, 2012
|
|
1I3U
 
 | | THREE-DIMENSIONAL STRUCTURE OF A LLAMA VHH DOMAIN COMPLEXED WITH THE DYE RR1 | | Descriptor: | 5-(4,6-DIAMINO-[1,3,5]TRIAZIN-2-YLAMINO)-4-HYDROXY-3-(2-SULFO-PHENYLAZO)-NAPHTHALENE-2,7-DISULFONIC ACID, ANTIBODY VHH LAMA DOMAIN, SULFATE ION | | Authors: | Spinelli, S, Tegoni, M, Frenken, L, van Vliet, C, Cambillau, C. | | Deposit date: | 2001-02-16 | | Release date: | 2001-08-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Lateral recognition of a dye hapten by a llama VHH domain.
J.Mol.Biol., 311, 2001
|
|
1I3V
 
 | | THREE-DIMENSIONAL STRUCTURE OF A LAMA VHH DOMAIN UNLIGANDED | | Descriptor: | ANTIBODY VHH LAMA DOMAIN | | Authors: | Spinelli, S, Tegoni, M, Frenken, L, van Vliet, C, Cambillau, C. | | Deposit date: | 2001-02-16 | | Release date: | 2001-08-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Lateral recognition of a dye hapten by a llama VHH domain.
J.Mol.Biol., 311, 2001
|
|
4RGA
 
 | | Phage 1358 receptor binding protein in complex with the trisaccharide GlcNAc-Galf-GlcOMe | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-beta-D-galactofuranose-(1-6)-methyl alpha-D-glucopyranoside, Phage 1358 receptor binding protein (ORF20) | | Authors: | Spinelli, S, Mccabe, O, Farenc, C, Tremblay, D, Blangy, S, Oscarson, S, Moineau, S, Cambillau, C. | | Deposit date: | 2014-09-29 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The targeted recognition of Lactococcus lactis phages to their polysaccharide receptors.
Mol.Microbiol., 96, 2015
|
|
2PVS
 
 | | Structure of human pancreatic lipase related protein 2 mutant N336Q | | Descriptor: | CALCIUM ION, Pancreatic lipase-related protein 2, SULFATE ION | | Authors: | Spinelli, S, Eydoux, C, Carriere, F, Cambillau, C. | | Deposit date: | 2007-05-10 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of human pancreatic lipase-related protein 2 with the lid in an open conformation.
Biochemistry, 47, 2008
|
|
