1BFI
 
 | | SOLUTION STRUCTURE OF THE C-TERMINAL SH2 DOMAIN OF THE P85ALPHA REGULATORY SUBUNIT OF PHOSPHOINOSITIDE 3-KINASE, NMR, 30 STRUCTURES | | Descriptor: | P85 ALPHA | | Authors: | Siegal, G, Davis, B, Kristensen, S.M, Sankar, A, Linacre, J, Stein, R.C, Panayotou, G, Waterfield, M.D, Driscoll, P.C. | | Deposit date: | 1997-11-18 | | Release date: | 1998-02-25 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal SH2 domain of the p85 alpha regulatory subunit of phosphoinositide 3-kinase.
J.Mol.Biol., 276, 1998
|
|
1BFJ
 
 | | SOLUTION STRUCTURE OF THE C-TERMINAL SH2 DOMAIN OF THE P85ALPHA REGULATORY SUBUNIT OF PHOSPHOINOSITIDE 3-KINASE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | P85 ALPHA | | Authors: | Siegal, G, Davis, B, Kristensen, S.M, Sankar, A, Linacre, J, Stein, R.C, Panayotou, G, Waterfield, M.D, Driscoll, P.C. | | Deposit date: | 1997-11-18 | | Release date: | 1998-02-25 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal SH2 domain of the p85 alpha regulatory subunit of phosphoinositide 3-kinase.
J.Mol.Biol., 276, 1998
|
|
1B64
 
 | | SOLUTION STRUCTURE OF THE GUANINE NUCLEOTIDE EXCHANGE FACTOR DOMAIN FROM HUMAN ELONGATION FACTOR-ONE BETA, NMR, 20 STRUCTURES | | Descriptor: | ELONGATION FACTOR 1-BETA | | Authors: | Perez, J.M.J, Siegal, G, Kriek, J, Hard, K, Dijk, J, Canters, G.W, Moller, W. | | Deposit date: | 1999-01-20 | | Release date: | 1999-05-18 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the guanine nucleotide exchange domain of human elongation factor 1beta reveals a striking resemblance to that of EF-Ts from Escherichia coli.
Structure Fold.Des., 7, 1999
|
|
2R63
 
 | | STRUCTURAL ROLE OF A BURIED SALT BRIDGE IN THE 434 REPRESSOR DNA-BINDING DOMAIN, NMR, 20 STRUCTURES | | Descriptor: | REPRESSOR PROTEIN FROM BACTERIOPHAGE 434 | | Authors: | Pervushin, K.V, Billeter, M, Siegal, G, Wuthrich, K. | | Deposit date: | 1996-11-13 | | Release date: | 1997-06-16 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Structural role of a buried salt bridge in the 434 repressor DNA-binding domain.
J.Mol.Biol., 264, 1996
|
|
1MO8
 
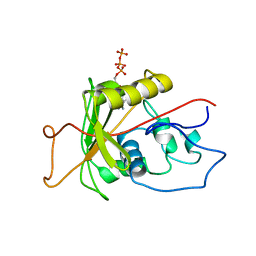 | | ATPase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Sodium/Potassium-Transporting ATPase alpha-1 | | Authors: | Hilge, M, Siegal, G, Vuister, G.W, Guentert, P, Gloor, S.M, Abrahams, J.P. | | Deposit date: | 2002-09-08 | | Release date: | 2003-06-10 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | ATP-induced conformational changes of the nucleotide-binding domain of Na,K-ATPase
Nat.Struct.Biol., 10, 2003
|
|
1MO7
 
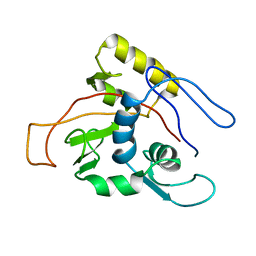 | | ATPase | | Descriptor: | Sodium/Potassium-transporting ATPase alpha-1 chain | | Authors: | Hilge, M, Siegal, G, Vuister, G.W, Guentert, P, Gloor, S.M, Abrahams, J.P. | | Deposit date: | 2002-09-08 | | Release date: | 2003-06-03 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | ATP-induced conformational changes of the nucleotide-binding domain of Na,K-ATPase
Nat.Struct.Biol., 10, 2003
|
|
2K7F
 
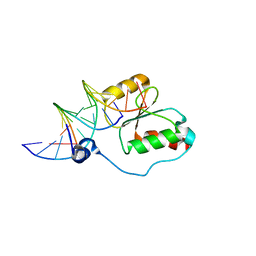 | | HADDOCK calculated model of the complex between the BRCT region of RFC p140 and dsDNA | | Descriptor: | 5'-D(P*DCP*DGP*DAP*DCP*DCP*DTP*DCP*DGP*DAP*DGP*DAP*DTP*DCP*DA)-3', 5'-D(P*DCP*DTP*DCP*DGP*DAP*DGP*DGP*DTP*DCP*DG)-3', Replication factor C subunit 1 | | Authors: | Kobayashi, M, Ab, E, Bonvin, A, Siegal, G. | | Deposit date: | 2008-08-10 | | Release date: | 2009-09-08 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure of the DNA-bound BRCA1 C-terminal region from human replication factor C p140 and model of the protein-DNA complex.
J.Biol.Chem., 285, 2010
|
|
2K6G
 
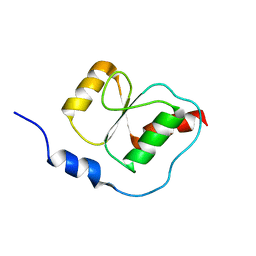 | |
1PBU
 
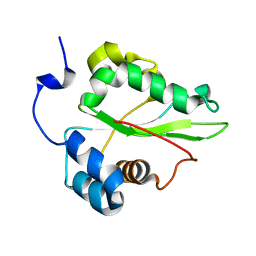 | | Solution structure of the C-terminal domain of the human eEF1Bgamma subunit | | Descriptor: | Elongation factor 1-gamma | | Authors: | Vanwetswinkel, S, Kriek, J, Andersen, G.R, Guntert, P, Dijk, J, Canters, G.W, Siegal, G. | | Deposit date: | 2003-05-15 | | Release date: | 2003-07-15 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | 1H, (15)N and (13)C resonance assignments of the highly conserved 19 kDa C-terminal domain from human Elongation Factor 1Bgamma.
J.Biomol.Nmr, 26, 2003
|
|
1QAD
 
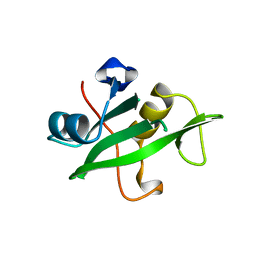 | | Crystal Structure of the C-Terminal SH2 Domain of the P85 alpha Regulatory Subunit of Phosphoinositide 3-Kinase: An SH2 domain mimicking its own substrate | | Descriptor: | PI3-KINASE P85 ALPHA SUBUNIT | | Authors: | Hoedemaeker, P.J, Siegal, G, Roe, M, Driscoll, P.C, Abrahams, J.P.A. | | Deposit date: | 1999-02-26 | | Release date: | 1999-10-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the C-terminal SH2 domain of the p85alpha regulatory subunit of phosphoinositide 3-kinase: an SH2 domain mimicking its own substrate.
J.Mol.Biol., 292, 1999
|
|
1R63
 
 | | STRUCTURAL ROLE OF A BURIED SALT BRIDGE IN THE 434 REPRESSOR DNA-BINDING DOMAIN, NMR, 20 STRUCTURES | | Descriptor: | REPRESSOR PROTEIN FROM BACTERIOPHAGE 434 | | Authors: | Pervushin, K.V, Billeter, M, Siegal, G, Wuthrich, K. | | Deposit date: | 1996-11-08 | | Release date: | 1997-06-16 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structural role of a buried salt bridge in the 434 repressor DNA-binding domain.
J.Mol.Biol., 264, 1996
|
|
2XYU
 
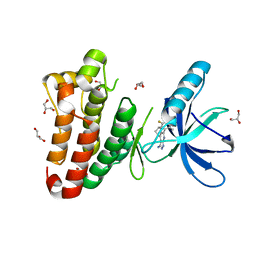 | | Crystal structure of EphA4 kinase domain in complex with VUF 12058 | | Descriptor: | 5-(5-FLUORO-2-METHYLPHENYL)-6,7,8,9-TETRAHYDRO-3H-PYRAZOLO[3,4-C]ISOQUINOLIN-1-AMINE, EPHRIN TYPE-A RECEPTOR 4,, GLYCEROL | | Authors: | Farenc, C.J.A, Celie, P.H.N, vanLinden, O.P.J, Siegal, G. | | Deposit date: | 2010-11-19 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.117 Å) | | Cite: | Fragment Based Lead Discovery of Small Molecule Inhibitors for the Epha4 Receptor Tyrosine Kinase.
Eur.J.Med.Chem., 47, 2012
|
|
2Y6O
 
 | |
2Y6M
 
 | |
4KLZ
 
 | | Inhibition of Small GTPases by Stabilization of the GDP Complex, a Novel Approach applied to Rit1, a Target for Rheumatoid Arthritis | | Descriptor: | GTP-binding protein Rit1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Shah, D.M, Kobayashi, M, Keizers, P.H, Tuin, A.W, Ab, E, Manning, L, Rzepiela, A.A, Andrews, M, Hoedemaeker, F.J, Siegal, G. | | Deposit date: | 2013-05-07 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Inhibition of Small GTPases by Stabilization of the GDP Complex, a Novel Approach applied to Rit1, a Target for Rheumatoid Arthritis
To be Published
|
|
4KZ6
 
 | | Crystal structure of AmpC beta-lactamase in complex with fragment 13 ((2R,6R)-6-methyl-1-(3-sulfanylpropanoyl)piperidine-2-carboxylic acid) | | Descriptor: | (2R,6R)-6-methyl-1-(3-sulfanylpropanoyl)piperidine-2-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Eidam, O, Barelier, S, Fish, I, Shoichet, B.K. | | Deposit date: | 2013-05-29 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Increasing chemical space coverage by combining empirical and computational fragment screens.
Acs Chem.Biol., 9, 2014
|
|
4KZA
 
 | | Crystal structure of AmpC beta-lactamase in complex with fragment 48 (3-(cyclopropylsulfamoyl)thiophene-2-carboxylic acid) | | Descriptor: | 3-(cyclopropylsulfamoyl)thiophene-2-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Eidam, O, Barelier, S, Fish, I, Shoichet, B.K. | | Deposit date: | 2013-05-29 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Increasing chemical space coverage by combining empirical and computational fragment screens.
Acs Chem.Biol., 9, 2014
|
|
4KZ7
 
 | | Crystal structure of AmpC beta-lactamase in complex with fragment 16 ((1R,4S)-4,7,7-trimethyl-3-oxo-2-oxabicyclo[2.2.1]heptane-1-carboxylic acid) | | Descriptor: | (1R,4S)-4,7,7-trimethyl-3-oxo-2-oxabicyclo[2.2.1]heptane-1-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Eidam, O, Barelier, S, Fish, I, Shoichet, B.K. | | Deposit date: | 2013-05-29 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Increasing chemical space coverage by combining empirical and computational fragment screens.
Acs Chem.Biol., 9, 2014
|
|
4KZ9
 
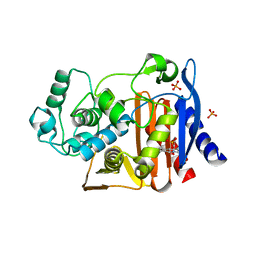 | | Crystal structure of AmpC beta-lactamase in complex with fragment 41 ((4R,4aS,8aS)-4-phenyldecahydroquinolin-4-ol) | | Descriptor: | (4R,4aS,8aS)-4-phenyldecahydroquinolin-4-ol, Beta-lactamase, PHOSPHATE ION | | Authors: | Eidam, O, Barelier, S, Fish, I, Shoichet, B.K. | | Deposit date: | 2013-05-29 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Increasing chemical space coverage by combining empirical and computational fragment screens.
Acs Chem.Biol., 9, 2014
|
|
4KZ5
 
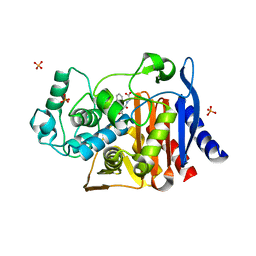 | | Crystal structure of AmpC beta-lactamase in complex with fragment 5 (N-{[3-(2-chlorophenyl)-5-methyl-1,2-oxazol-4-yl]carbonyl}glycine) | | Descriptor: | Beta-lactamase, N-{[3-(2-chlorophenyl)-5-methyl-1,2-oxazol-4-yl]carbonyl}glycine, PHOSPHATE ION | | Authors: | Eidam, O, Barelier, S, Fish, I, Shoichet, B.K. | | Deposit date: | 2013-05-29 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Increasing chemical space coverage by combining empirical and computational fragment screens.
Acs Chem.Biol., 9, 2014
|
|
4KZ3
 
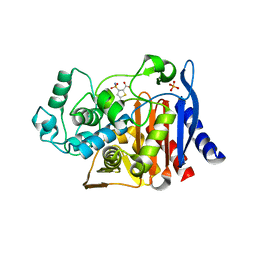 | | Crystal structure of AmpC beta-lactamase in complex with fragment 44 (5-chloro-3-sulfamoylthiophene-2-carboxylic acid) | | Descriptor: | 5-chloro-3-sulfamoylthiophene-2-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Eidam, O, Barelier, S, Fish, I, Shoichet, B.K. | | Deposit date: | 2013-05-29 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Increasing chemical space coverage by combining empirical and computational fragment screens.
Acs Chem.Biol., 9, 2014
|
|
4KZB
 
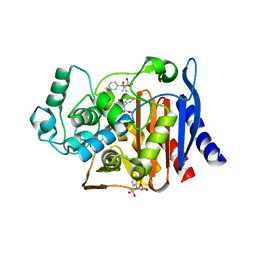 | | Crystal structure of AmpC beta-lactamase in complex with fragment 50 (N-(methylsulfonyl)-N-phenyl-alanine) | | Descriptor: | Beta-lactamase, N-(methylsulfonyl)-N-phenyl-D-alanine, N-(methylsulfonyl)-N-phenyl-L-alanine | | Authors: | Eidam, O, Barelier, S, Fish, I, Shoichet, B.K. | | Deposit date: | 2013-05-29 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Increasing chemical space coverage by combining empirical and computational fragment screens.
Acs Chem.Biol., 9, 2014
|
|
4KZ8
 
 | | Crystal structure of AmpC beta-lactamase in complex with fragment 20 (1,3-diethyl-2-thioxodihydropyrimidine-4,6(1H,5H)-dione) | | Descriptor: | 1,3-diethyl-2-thioxodihydropyrimidine-4,6(1H,5H)-dione, Beta-lactamase, PHOSPHATE ION | | Authors: | Eidam, O, Barelier, S, Fish, I, Shoichet, B.K. | | Deposit date: | 2013-05-29 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.282 Å) | | Cite: | Increasing chemical space coverage by combining empirical and computational fragment screens.
Acs Chem.Biol., 9, 2014
|
|
4KZ4
 
 | | Crystal structure of AmpC beta-lactamase in complex with fragment 60 (2-[(propylsulfonyl)amino]benzoic acid) | | Descriptor: | 2-[(propylsulfonyl)amino]benzoic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Eidam, O, Barelier, S, Fish, I, Shoichet, B.K. | | Deposit date: | 2013-05-29 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Increasing chemical space coverage by combining empirical and computational fragment screens.
Acs Chem.Biol., 9, 2014
|
|
5FE2
 
 | | Crystal structure of human PCAF bromodomain in complex with fragment BR013 (fragment 3) | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-3~{H}-isoindol-1-one, Histone acetyltransferase KAT2B | | Authors: | Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Based Identification of Inhibitory Fragments Targeting the p300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 59, 2016
|
|
