3CDJ
 
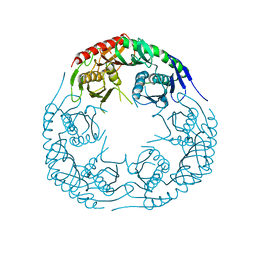 | | Crystal structure of the E. coli KH/S1 domain truncated PNPase | | Descriptor: | Polynucleotide phosphorylase | | Authors: | Shi, Z, Yang, W.Z, Lin-Chao, S, Chak, K.F, Yuan, H.S. | | Deposit date: | 2008-02-27 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Escherichia coli PNPase: central channel residues are involved in processive RNA degradation.
Rna, 14, 2008
|
|
3CDI
 
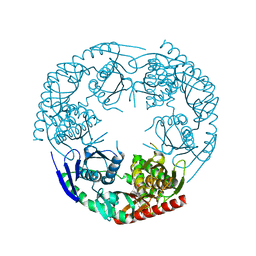 | | Crystal structure of E. coli PNPase | | Descriptor: | Polynucleotide phosphorylase | | Authors: | Shi, Z, Yang, W.Z, Lin-Chao, S, Chak, K.F, Yuan, H.S. | | Deposit date: | 2008-02-27 | | Release date: | 2008-12-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Escherichia coli PNPase: central channel residues are involved in processive RNA degradation.
Rna, 14, 2008
|
|
1ZNS
 
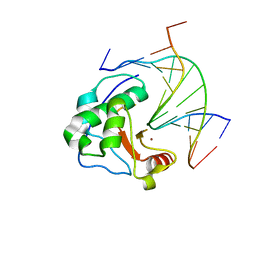 | | Crystal structure of N-ColE7/12-bp DNA/Zn complex | | Descriptor: | 5'-D(*CP*GP*GP*GP*AP*TP*AP*TP*CP*CP*CP*G)-3', Colicin E7, ZINC ION | | Authors: | Doudeva, L.G, Huang, H, Hsia, K.C, Shi, Z, Li, C.L, Shen, Y, Yuan, H.S. | | Deposit date: | 2005-05-12 | | Release date: | 2006-03-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structural analysis and metal-dependent stability and activity studies of the ColE7 endonuclease domain in complex with DNA/Zn2+ or inhibitor/Ni2+
Protein Sci., 15, 2006
|
|
1ZNV
 
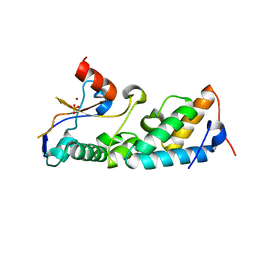 | | How a His-metal finger endonuclease ColE7 binds and cleaves DNA with a transition metal ion cofactor | | Descriptor: | Colicin E7, Colicin E7 immunity protein, NICKEL (II) ION, ... | | Authors: | Doudeva, L.G, Huang, H, Hsia, K.C, Shi, Z, Li, C.L, Shen, Y, Yuan, H.S. | | Deposit date: | 2005-05-12 | | Release date: | 2006-03-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structural analysis and metal-dependent stability and activity studies of the ColE7 endonuclease domain in complex with DNA/Zn2+ or inhibitor/Ni2+
Protein Sci., 15, 2006
|
|
2AXC
 
 | | Crystal structure of ColE7 translocation domain | | Descriptor: | Colicin E7, GLYCEROL, SULFATE ION | | Authors: | Cheng, Y.S, Shi, Z, Doudeva, L.G, Yang, W.Z, Chak, K.F, Yuan, H.S. | | Deposit date: | 2005-09-04 | | Release date: | 2006-03-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-resolution crystal structure of a truncated ColE7 translocation domain: implications for colicin transport across membranes
J.Mol.Biol., 356, 2006
|
|
4LN0
 
 | | Crystal structure of the VGLL4-TEAD4 complex | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Transcription cofactor vestigial-like protein 4, ... | | Authors: | Wang, H, Shi, Z, Zhou, Z. | | Deposit date: | 2013-07-11 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.896 Å) | | Cite: | A Peptide Mimicking VGLL4 Function Acts as a YAP Antagonist Therapy against Gastric Cancer.
Cancer Cell, 25, 2014
|
|
4N6J
 
 | | Crystal structure of human Striatin-3 coiled coil domain | | Descriptor: | Striatin-3 | | Authors: | Chen, C, Shi, Z, Zhou, Z. | | Deposit date: | 2013-10-13 | | Release date: | 2014-02-26 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Striatins contain a noncanonical coiled coil that binds protein phosphatase 2A A subunit to form a 2:2 heterotetrameric core of striatin-interacting phosphatase and kinase (STRIPAK) complex.
J.Biol.Chem., 289, 2014
|
|
7VCS
 
 | | Human p97 double hexamer conformer II with ATPgammaS bound | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Gao, H, Li, F, Shi, Z, Li, Y, Yu, H. | | Deposit date: | 2021-09-03 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-09 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Cryo-EM structures of human p97 double hexamer capture potentiated ATPase-competent state.
Cell Discov, 8, 2022
|
|
7VCX
 
 | | Human p97 single hexamer conformer II with ATPgammaS bound | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Gao, H, Li, F, Shi, Z, Li, Y, Yu, H. | | Deposit date: | 2021-09-04 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-09 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Cryo-EM structures of human p97 double hexamer capture potentiated ATPase-competent state.
Cell Discov, 8, 2022
|
|
7VCT
 
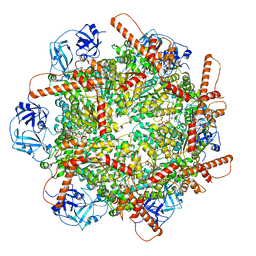 | | Human p97 single hexamer conformer III with D1-ATPgammaS and D2-ADP bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gao, H, Li, F, Shi, Z, Li, Y, Yu, H. | | Deposit date: | 2021-09-04 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-09 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Cryo-EM structures of human p97 double hexamer capture potentiated ATPase-competent state.
Cell Discov, 8, 2022
|
|
7VCV
 
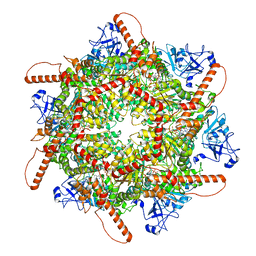 | | Human p97 single hexamer conformer I with ATPgammaS bound | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Gao, H, Li, F, Shi, Z, Li, Y, Yu, H. | | Deposit date: | 2021-09-04 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-09 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Cryo-EM structures of human p97 double hexamer capture potentiated ATPase-competent state.
Cell Discov, 8, 2022
|
|
7VCU
 
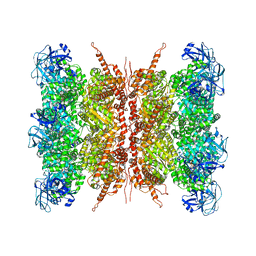 | | Human p97 double hexamer conformer I with D1-ATPgammaS and D2-ADP bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gao, H, Li, F, Shi, Z, Li, Y, Yu, H. | | Deposit date: | 2021-09-04 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-09 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Cryo-EM structures of human p97 double hexamer capture potentiated ATPase-competent state.
Cell Discov, 8, 2022
|
|
3UD8
 
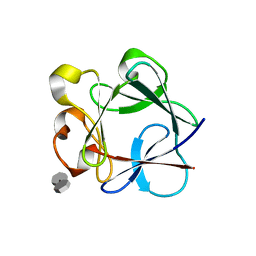 | | Crystal Structure Analysis of FGF1-Disaccharide(NI22) complex | | Descriptor: | 2-deoxy-3-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-1-O-methyl-2-O-sulfo-alpha-L-idopyranuronic acid, Heparin-binding growth factor 1, PHOSPHATE ION | | Authors: | Hung, S.-C, Shi, Z. | | Deposit date: | 2011-10-27 | | Release date: | 2012-11-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Divergent synthesis of 48 heparan sulfate-based disaccharides and probing the specific sugar-fibroblast growth factor-1 interaction
J.Am.Chem.Soc., 134, 2012
|
|
3UDA
 
 | | Crystal Structure Analysis of FGF1-Disaccharide(NI24) complex | | Descriptor: | 2-deoxy-3,6-di-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-1-O-methyl-2-O-sulfo-alpha-L-idopyranuronic acid, Heparin-binding growth factor 1, PHOSPHATE ION | | Authors: | Hung, S.-C, Shi, Z. | | Deposit date: | 2011-10-27 | | Release date: | 2012-11-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Divergent synthesis of 48 heparan sulfate-based disaccharides and probing the specific sugar-fibroblast growth factor-1 interaction
J.Am.Chem.Soc., 134, 2012
|
|
3UD9
 
 | | Crystal Structure Analysis of FGF1-Disaccharide(NI23) complex | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-1-O-methyl-2-O-sulfo-alpha-L-idopyranuronic acid, Heparin-binding growth factor 1, PHOSPHATE ION | | Authors: | Hung, S.-C, Shi, Z. | | Deposit date: | 2011-10-27 | | Release date: | 2012-11-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Divergent synthesis of 48 heparan sulfate-based disaccharides and probing the specific sugar-fibroblast growth factor-1 interaction
J.Am.Chem.Soc., 134, 2012
|
|
3UD7
 
 | | Crystal Structure Analysis of FGF1-Disaccharide(NI21) complexes | | Descriptor: | 2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-1-O-methyl-2-O-sulfo-alpha-L-idopyranuronic acid, Heparin-binding growth factor 1, PHOSPHATE ION | | Authors: | Hung, S.-C, Shi, Z. | | Deposit date: | 2011-10-27 | | Release date: | 2012-11-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Divergent synthesis of 48 heparan sulfate-based disaccharides and probing the specific sugar-fibroblast growth factor-1 interaction
J.Am.Chem.Soc., 134, 2012
|
|
4Z8M
 
 | | Crystal structure of the MAVS-TRAF6 complex | | Descriptor: | Peptide from Mitochondrial antiviral-signaling protein, TNF receptor-associated factor 6 | | Authors: | Shi, Z.B, Zhou, Z. | | Deposit date: | 2015-04-09 | | Release date: | 2015-09-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural Insights into Mitochondrial Antiviral Signaling Protein (MAVS)-Tumor Necrosis Factor Receptor-associated Factor 6 (TRAF6) Signaling
J.Biol.Chem., 290, 2015
|
|
4FZA
 
 | | Crystal structure of MST4-MO25 complex | | Descriptor: | Calcium-binding protein 39, GLYCEROL, Serine/threonine-protein kinase MST4 | | Authors: | Shi, Z.B, Zhou, Z.C. | | Deposit date: | 2012-07-06 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structure of the MST4 in Complex with MO25 Provides Insights into Its Activation Mechanism
Structure, 21, 2013
|
|
4FZF
 
 | | Crystal structure of MST4-MO25 complex with DKI | | Descriptor: | 5-AMINO-3-{[4-(AMINOSULFONYL)PHENYL]AMINO}-N-(2,6-DIFLUOROPHENYL)-1H-1,2,4-TRIAZOLE-1-CARBOTHIOAMIDE, Calcium-binding protein 39, Serine/threonine-protein kinase MST4 | | Authors: | Shi, Z.B, Zhou, Z.C. | | Deposit date: | 2012-07-06 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.64 Å) | | Cite: | Structure of the MST4 in Complex with MO25 Provides Insights into Its Activation Mechanism
Structure, 21, 2013
|
|
4FZD
 
 | | Crystal structure of MST4-MO25 complex with WSF motif | | Descriptor: | C-terminal peptide from Serine/threonine-protein kinase MST4, Calcium-binding protein 39, GLYCEROL, ... | | Authors: | Shi, Z.B, Zhou, Z.C. | | Deposit date: | 2012-07-06 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure of the MST4 in Complex with MO25 Provides Insights into Its Activation Mechanism
Structure, 21, 2013
|
|
6WG3
 
 | | Cryo-EM structure of human Cohesin-NIPBL-DNA complex | | Descriptor: | Cohesin subunit SA-1, DNA (51-MER), Double-strand-break repair protein rad21 homolog, ... | | Authors: | Shi, Z.B, Gao, H, Bai, X.C, Yu, H. | | Deposit date: | 2020-04-04 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Cryo-EM structure of the human cohesin-NIPBL-DNA complex.
Science, 368, 2020
|
|
6WG4
 
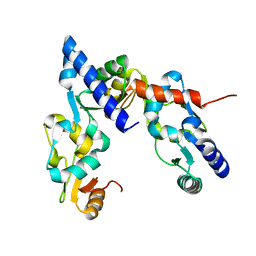 | |
6WGE
 
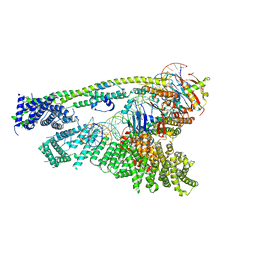 | | Cryo-EM structure of human Cohesin-NIPBL-DNA complex without STAG1 | | Descriptor: | DNA (43-MER), Double-strand-break repair protein rad21 homolog, MAGNESIUM ION, ... | | Authors: | Shi, Z.B, Gao, H, Bai, X.C, Yu, H. | | Deposit date: | 2020-04-05 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of the human cohesin-NIPBL-DNA complex.
Science, 368, 2020
|
|
6WG6
 
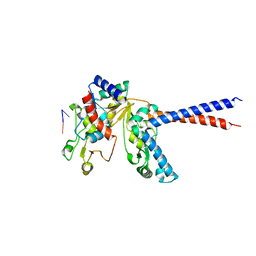 | |
5XY9
 
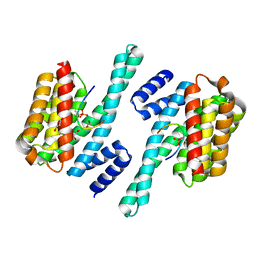 | | Structure of the MST4 and 14-3-3 complex | | Descriptor: | 14-3-3 protein zeta/delta, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, GLYCEROL, ... | | Authors: | Shi, Z.B, Zhou, Z.C. | | Deposit date: | 2017-07-06 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Structure of the MST4 and 14-3-3 complex
To Be Published
|
|
