5K31
 
 | | Crystal structure of Human fibrillar procollagen type I C-propeptide Homo-trimer | | Descriptor: | CALCIUM ION, CHLORIDE ION, Collagen alpha-1(I) chain, ... | | Authors: | Sharma, U, Hulmes, D.J.S, Aghajari, N. | | Deposit date: | 2016-05-19 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of homo- and heterotrimerization of collagen I.
Nat Commun, 8, 2017
|
|
7EL5
 
 | |
6LH6
 
 | |
3P8S
 
 | |
3S18
 
 | |
3V6N
 
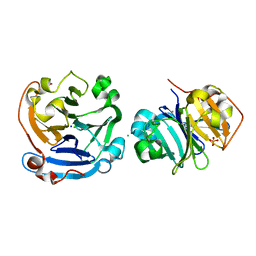 | |
6XVP
 
 | | Crystal structure of Neprilysin in complex with Sampatrilat. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Neprilysin, ... | | Authors: | Cozier, G.E, Acharya, K.R, Sharma, U. | | Deposit date: | 2020-01-22 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Molecular Basis for Omapatrilat and Sampatrilat Binding to Neprilysin-Implications for Dual Inhibitor Design with Angiotensin-Converting Enzyme.
J.Med.Chem., 63, 2020
|
|
6SVY
 
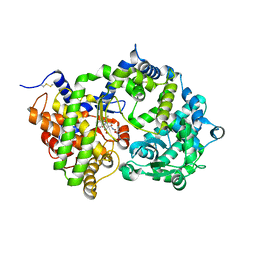 | | Crystal structure of Neprilysin in complex with Sampatrilat-ASP. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Neprilysin, Sampatrilat-Asp, ... | | Authors: | Cozier, G.E, Acharya, K.R, Sharma, U. | | Deposit date: | 2019-09-19 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Basis for Omapatrilat and Sampatrilat Binding to Neprilysin-Implications for Dual Inhibitor Design with Angiotensin-Converting Enzyme.
J.Med.Chem., 63, 2020
|
|
6SUK
 
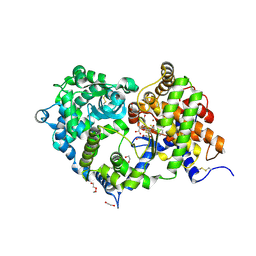 | | Crystal structure of Neprilysin in complex with Omapatrilat. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Cozier, G.E, Acharya, K.R, Sharma, U. | | Deposit date: | 2019-09-15 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular Basis for Omapatrilat and Sampatrilat Binding to Neprilysin-Implications for Dual Inhibitor Design with Angiotensin-Converting Enzyme.
J.Med.Chem., 63, 2020
|
|
1SY8
 
 | | Structure of DNA sequence d-TGATCA by two-dimensional nuclear magnetic resonance spec and restrained molecular dynamics | | Descriptor: | 5'-D(P*TP*GP*AP*TP*CP*A)-3' | | Authors: | Barthwal, R, Awasthi, P, Narang, M, Sharma, U, Srivastava, N. | | Deposit date: | 2004-04-01 | | Release date: | 2005-01-04 | | Last modified: | 2012-02-22 | | Method: | SOLUTION NMR | | Cite: | Structure of DNA sequence d-TGATCA by two-dimensional nuclear magnetic resonance spectroscopy and restrained molecular dynamics
J.STRUCT.BIOL., 148, 2004
|
|
2K84
 
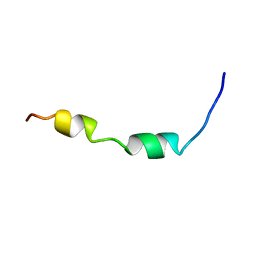 | | Solution Structure of the equine infectious anemia virus p9 GAG protein | | Descriptor: | P9 | | Authors: | Sharma, A, Bruns, K, Roeder, R, Henklein, P, Votteler, J, Wray, V, Sharma, U. | | Deposit date: | 2008-09-02 | | Release date: | 2009-09-08 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Equine Infectious Anemia Virus p9 protein: a rationalization of its different ALIX binding requirements compared to the analogous HIV-p6 protein
Bmc Struct.Biol., 9, 2009
|
|
6FZW
 
 | |
6FZV
 
 | |
