4UQT
 
 | | RRM-peptide structure in RES complex | | Descriptor: | PRE-MRNA-SPLICING FACTOR CWC26, U2 SNRNP COMPONENT IST3 | | Authors: | Tripsianes, K, Friberg, A, Barrandon, C, Seraphin, B, Sattler, M. | | Deposit date: | 2014-06-25 | | Release date: | 2014-09-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A Novel Protein-Protein Interaction in the Res (Retention and Splicing) Complex.
J.Biol.Chem., 289, 2014
|
|
4X33
 
 | | Structure of the Elongator cofactor complex Kti11/Kti13 at 1.45A | | Descriptor: | 1,2-DIMETHOXYETHANE, CHLORIDE ION, Diphthamide biosynthesis protein 3, ... | | Authors: | Kolaj-Robin, O, McEwen, A.G, Cavarelli, J, Seraphin, B. | | Deposit date: | 2014-11-27 | | Release date: | 2015-01-21 | | Last modified: | 2015-03-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of the Elongator cofactor complex Kti11/Kti13 provides insight into the role of Kti13 in Elongator-dependent tRNA modification.
Febs J., 282, 2015
|
|
1I5L
 
 | | CRYSTAL STRUCTURE OF AN SM-LIKE PROTEIN (AF-SM1) FROM ARCHAEOGLOBUS FULGIDUS COMPLEXED WITH SHORT POLY-U RNA | | Descriptor: | 5'-R(*UP*UP*U)-3', PUTATIVE SNRNP SM-LIKE PROTEIN AF-SM1, URIDINE | | Authors: | Toro, I, Thore, S, Mayer, C, Basquin, J, Seraphin, B, Suck, D. | | Deposit date: | 2001-02-28 | | Release date: | 2001-08-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | RNA binding in an Sm core domain: X-ray structure and functional analysis of an archaeal Sm protein complex.
EMBO J., 20, 2001
|
|
2FHO
 
 | | NMR solution structure of the human spliceosomal protein complex p14-SF3b155 | | Descriptor: | spliceosomal protein SF3b155, spliceosomal protein p14 | | Authors: | Kuwasako, K, Dohmae, N, Inoue, M, Shirouzu, M, Guntert, P, Seraphin, B, Muto, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-26 | | Release date: | 2006-12-26 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the human spliceosomal protein complex p14-SF3b155
To be Published
|
|
1I4K
 
 | | CRYSTAL STRUCTURE OF AN SM-LIKE PROTEIN (AF-SM1) FROM ARCHAEOGLOBUS FULGIDUS AT 2.5A RESOLUTION | | Descriptor: | CITRIC ACID, PUTATIVE SNRNP SM-LIKE PROTEIN | | Authors: | Toro, I, Thore, S, Mayer, C, Basquin, J, Seraphin, B, Suck, D. | | Deposit date: | 2001-02-22 | | Release date: | 2001-08-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | RNA binding in an Sm core domain: X-ray structure and functional analysis of an archaeal Sm protein complex.
EMBO J., 20, 2001
|
|
3MGU
 
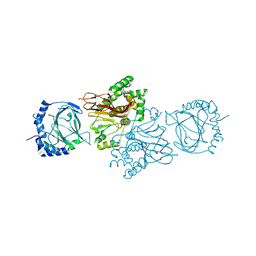 | | Structure of S. cerevisiae Tpa1 protein, a proline hydroxylase modifying ribosomal protein Rps23 | | Descriptor: | FE (II) ION, PKHD-type hydroxylase TPA1 | | Authors: | Henri, J, Rispal, D, Bayart, E, van Tilbeurgh, H, Seraphin, B, Graille, M. | | Deposit date: | 2010-04-07 | | Release date: | 2010-07-14 | | Last modified: | 2018-01-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and functional insights into S. cerevisiae Tpa1, a putative prolyl hydroxylase influencing translation termination and transcription
To be Published
|
|
4A9A
 
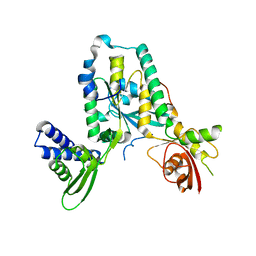 | | Structure of Rbg1 in complex with Tma46 dfrp domain | | Descriptor: | RIBOSOME-INTERACTING GTPASE 1, TRANSLATION MACHINERY-ASSOCIATED PROTEIN 46 | | Authors: | Francis, S.M, Gas, M, Daugeron, M, Seraphin, B, Bravo, J. | | Deposit date: | 2011-11-25 | | Release date: | 2012-10-03 | | Last modified: | 2017-01-25 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Rbg1-Tma46 Dimer Structure Reveals New Functional Domains and Their Role in Polysome Recruitment.
Nucleic Acids Res., 40, 2012
|
|
1UOC
 
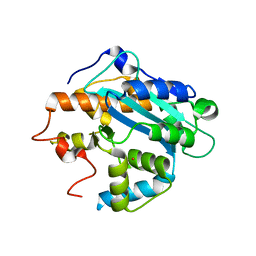 | | X-ray structure of the RNase domain of the yeast Pop2 protein | | Descriptor: | CALCIUM ION, POP2, XENON | | Authors: | Thore, S, Mauxion, F, Seraphin, B, Suck, D. | | Deposit date: | 2003-09-16 | | Release date: | 2003-11-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-Ray Structure and Activity of the Yeast Pop2 Protein: A Nuclease Subunit of the Mrna Deadenylase Complex
Embo Rep., 4, 2003
|
|
4OJJ
 
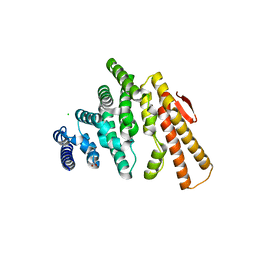 | | Structure of C-terminal domain from S. cerevisiae Pat1 decapping activator (Space group : P212121) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA topoisomerase 2-associated protein PAT1, ... | | Authors: | Fourati-Kammoun, Z, Kolesnikova, O, Back, R, Keller, J, Lazar, N, Gaudon-Plesse, C, Seraphin, B, Graille, M. | | Deposit date: | 2014-01-21 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | The C-terminal domain from S. cerevisiae Pat1 displays two conserved regions involved in decapping factor recruitment.
Plos One, 9, 2014
|
|
4OGP
 
 | | Structure of C-terminal domain from S. cerevisiae Pat1 decapping activator (Space group : P21) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA topoisomerase 2-associated protein PAT1 | | Authors: | Fourati-Kammoun, Z, Kolesnikova, O, Back, R, Keller, J, Lazar, N, Gaudon-Plesse, C, Seraphin, B, Graille, M. | | Deposit date: | 2014-01-16 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The C-terminal domain from S. cerevisiae Pat1 displays two conserved regions involved in decapping factor recruitment.
Plos One, 9, 2014
|
|
2MZS
 
 | |
2MZQ
 
 | |
2MZR
 
 | |
2MZT
 
 | |
3QTM
 
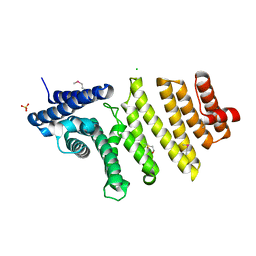 | | Structure of S. pombe nuclear import adaptor Nro1 (Space group P21) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Rispal, D, Henri, J, van Tilbeurgh, H, Graille, M, Seraphin, B. | | Deposit date: | 2011-02-23 | | Release date: | 2011-09-21 | | Last modified: | 2018-01-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and functional analysis of Nro1/Ett1: a protein involved in translation termination in S. cerevisiae and in O2-mediated gene control in S. pombe
Rna, 17, 2011
|
|
3QTN
 
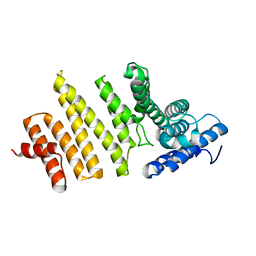 | | Structure of S. pombe nuclear import adaptor Nro1 (Space group P6522) | | Descriptor: | Uncharacterized protein C4B3.07 | | Authors: | Rispal, D, Henri, J, van Tilbeurgh, H, Graille, M, Seraphin, B. | | Deposit date: | 2011-02-23 | | Release date: | 2011-09-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.499 Å) | | Cite: | Structural and functional analysis of Nro1/Ett1: a protein involved in translation termination in S. cerevisiae and in O2-mediated gene control in S. pombe
Rna, 17, 2011
|
|
5LM5
 
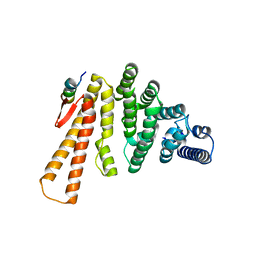 | | Structure of C-terminal domain from S. cerevisiae Pat1 decapping activator bound to Dcp2 HLM2 peptide (region 435-451) | | Descriptor: | DNA topoisomerase 2-associated protein PAT1, mRNA decapping protein 2 | | Authors: | Charenton, C, Gaudon-Plesse, C, Fourati, Z, Taverniti, V, Back, R, Kolesnikova, O, Seraphin, B, Graille, M. | | Deposit date: | 2016-07-29 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A unique surface on Pat1 C-terminal domain directly interacts with Dcp2 decapping enzyme and Xrn1 5'-3' mRNA exonuclease in yeast.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5LMG
 
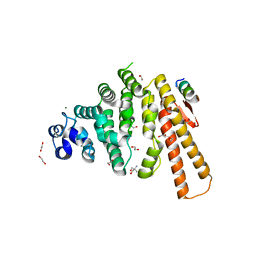 | | Structure of C-terminal domain from S. cerevisiae Pat1 decapping activator bound to Dcp2 HLM10 peptide (region 954-970) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA topoisomerase 2-associated protein PAT1, ... | | Authors: | Charenton, C, Gaudon-Plesse, C, Fourati, Z, Taverniti, V, Back, R, Kolesnikova, O, Seraphin, B, Graille, M. | | Deposit date: | 2016-07-30 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A unique surface on Pat1 C-terminal domain directly interacts with Dcp2 decapping enzyme and Xrn1 5'-3' mRNA exonuclease in yeast.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5LOP
 
 | | Structure of the active form of /K. lactis/ Dcp1-Dcp2-Edc3 decapping complex bound to m7GDP | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, KLLA0A11308p, KLLA0E01827p, ... | | Authors: | Charenton, C, Taverniti, V, Gaudon-Plesse, C, Back, R, Seraphin, B, Graille, M. | | Deposit date: | 2016-08-09 | | Release date: | 2016-10-05 | | Last modified: | 2017-09-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the active form of Dcp1-Dcp2 decapping enzyme bound to m(7)GDP and its Edc3 activator.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5LMF
 
 | | Structure of C-terminal domain from S. cerevisiae Pat1 decapping activator bound to Dcp2 HLM3 peptide (region 484-500) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA topoisomerase 2-associated protein PAT1, ... | | Authors: | Charenton, C, Gaudon-Plesse, C, Fourati, Z, Taverniti, V, Back, R, Kolesnikova, O, Seraphin, B, Graille, M. | | Deposit date: | 2016-07-30 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A unique surface on Pat1 C-terminal domain directly interacts with Dcp2 decapping enzyme and Xrn1 5'-3' mRNA exonuclease in yeast.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5LON
 
 | | Structure of /K. lactis/ Dcp1-Dcp2 decapping complex. | | Descriptor: | KLLA0E01827p, KLLA0F23980p | | Authors: | Charenton, C, Taverniti, V, Gaudon-Plesse, C, Back, R, Seraphin, B, Graille, M. | | Deposit date: | 2016-08-09 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the active form of Dcp1-Dcp2 decapping enzyme bound to m(7)GDP and its Edc3 activator.
Nat.Struct.Mol.Biol., 23, 2016
|
|
2JKD
 
 | | Structure of the yeast Pml1 splicing factor and its integration into the RES complex | | Descriptor: | GLYCEROL, PRE-MRNA LEAKAGE PROTEIN 1, SULFATE ION | | Authors: | Brooks, M.A, Dziembowski, A, Quevillon-Cheruel, S, Henriot, V, Faux, C, van Tilbeurgh, H, Seraphin, B. | | Deposit date: | 2008-08-27 | | Release date: | 2008-09-26 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Yeast Pml1 Splicing Factor and its Integration Into the Res Complex
Nucleic Acids Res., 37, 2009
|
|
6Y3Z
 
 | |
6Y3P
 
 | |
5L7J
 
 | |
