2XDF
 
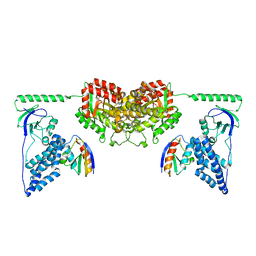 | | Solution Structure of the Enzyme I Dimer Complexed with HPr Using Residual Dipolar Couplings and Small Angle X-Ray Scattering | | Descriptor: | PHOSPHOCARRIER PROTEIN HPR, PHOSPHOENOLPYRUVATE-PROTEIN PHOSPHOTRANSFERASE | | Authors: | Schwieters, C.D, Suh, J.-Y, Grishaev, A, Guirlando, R, Takayama, Y, Clore, G.M. | | Deposit date: | 2010-04-30 | | Release date: | 2010-09-22 | | Last modified: | 2019-08-21 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Solution Structure of the 128 kDa Enzyme I Dimer from Escherichia Coli and its 146 kDa Complex with Hpr Using Residual Dipolar Couplings and Small- and Wide-Angle X-Ray Scattering.
J.Am.Chem.Soc., 132, 2010
|
|
2KX9
 
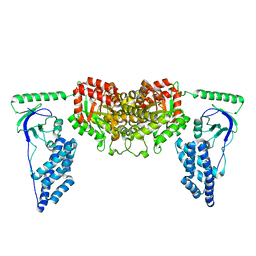 | | Solution Structure of the Enzyme I dimer Using Residual Dipolar Couplings and Small Angle X-Ray Scattering | | Descriptor: | Phosphoenolpyruvate-protein phosphotransferase | | Authors: | Schwieters, C.D, Suh, J, Grishaev, A, Takayama, Y, Guirlando, R, Clore, G. | | Deposit date: | 2010-04-29 | | Release date: | 2010-09-15 | | Last modified: | 2019-05-08 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Solution structure of the 128 kDa enzyme I dimer from Escherichia coli and its 146 kDa complex with HPr using residual dipolar couplings and small- and wide-angle X-ray scattering.
J.Am.Chem.Soc., 132, 2010
|
|
6N8C
 
 | | Structure of the Huntingtin tetramer/dimer mixture determined by paramagnetic NMR | | Descriptor: | Huntingtin | | Authors: | Schwieters, C.D, Kotler, S.A, Schmidt, T, Ceccon, A, Ghirlando, R, Libich, D.S, Clore, G.M. | | Deposit date: | 2018-11-29 | | Release date: | 2019-02-13 | | Last modified: | 2020-01-01 | | Method: | SOLUTION NMR | | Cite: | Probing initial transient oligomerization events facilitating Huntingtin fibril nucleation at atomic resolution by relaxation-based NMR.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
2XKM
 
 | | Consensus structure of Pf1 filamentous bacteriophage from X-ray fibre diffraction and solid-state NMR | | Descriptor: | CAPSID PROTEIN G8P | | Authors: | Straus, S.K, P Scott, W.R, Schwieters, C.D, Marvin, D.A. | | Deposit date: | 2010-07-09 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-20 | | Method: | FIBER DIFFRACTION (3.3 Å), SOLID-STATE NMR | | Cite: | Consensus Structure of Pf1 Filamentous Bacteriophage from X-Ray Fibre Diffraction and Solid-State NMR.
Eur.Biophys.J., 40, 2011
|
|
6X63
 
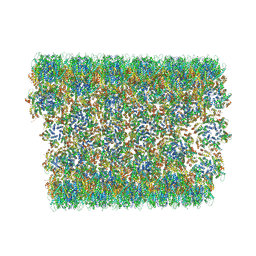 | | Atomic-Resolution Structure of HIV-1 Capsid Tubes by Magic Angle Spinning NMR | | Descriptor: | HIV-1 capsid protein | | Authors: | Lu, M, Russell, R.W, Bryer, A, Quinn, C.M, Hou, G, Zhang, H, Schwieters, C.D, Perilla, J.R, Gronenborn, A.M, Polenova, T. | | Deposit date: | 2020-05-27 | | Release date: | 2020-09-02 | | Last modified: | 2023-06-14 | | Method: | SOLID-STATE NMR | | Cite: | Atomic-resolution structure of HIV-1 capsid tubes by magic-angle spinning NMR.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8FPT
 
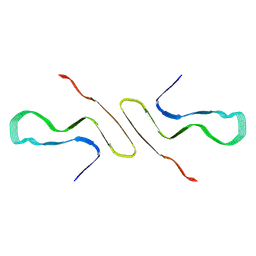 | | STRUCTURE OF ALPHA-SYNUCLEIN FIBRILS DERIVED FROM HUMAN LEWY BODY DEMENTIA TISSUE | | Descriptor: | Alpha-synuclein | | Authors: | Barclay, A.M, Dhavale, D.D, Borcik, C.G, Rau, M.J, Basore, K, Milchberg, M.H, Warmuth, O.A, Kotzbauer, P.T, Rienstra, C.M, Schwieters, C.D. | | Deposit date: | 2023-01-05 | | Release date: | 2023-02-22 | | Method: | SOLID-STATE NMR | | Cite: | Structure of alpha-synuclein fibrils derived from human Lewy body dementia tissue.
Biorxiv, 2023
|
|
7TA8
 
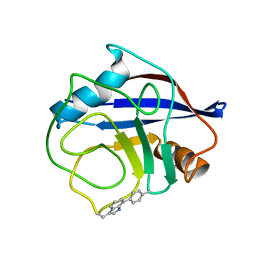 | | NMR structure of crosslinked cyclophilin A | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Lu, M, Toptygin, D, Xiang, Y, Shi, Y, Schwieters, C.D, Lipinski, E.C, Ahn, J, Byeon, I.-J.L, Gronenborn, A.M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-06-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Magic of Linking Rings: Discovery of a Unique Photoinduced Fluorescent Protein Crosslink.
J.Am.Chem.Soc., 144, 2022
|
|
6F3K
 
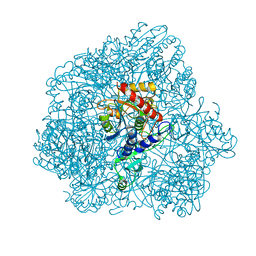 | | Combined solid-state NMR, solution-state NMR and EM data for structure determination of the tetrahedral aminopeptidase TET2 from P. horikoshii | | Descriptor: | Tetrahedral aminopeptidase, ZINC ION | | Authors: | Gauto, D.F, Estrozi, L.F, Schwieters, C.D, Effantin, G, Macek, P, Sounier, R, Kerfah, R, Sivertsen, A.C, Colletier, J.P, Boisbouvier, J, Schoehn, G, Favier, A, Schanda, P. | | Deposit date: | 2017-11-28 | | Release date: | 2018-03-14 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å), SOLID-STATE NMR, SOLUTION NMR | | Cite: | Integrated NMR and cryo-EM atomic-resolution structure determination of a half-megadalton enzyme complex.
Nat Commun, 10, 2019
|
|
5I1R
 
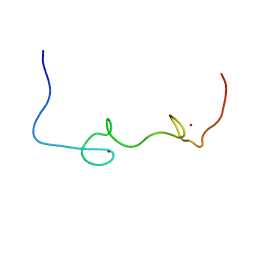 | | Quantitative characterization of configurational space sampled by HIV-1 nucleocapsid using solution NMR and X-ray scattering | | Descriptor: | Nucleocapsid protein p7, ZINC ION | | Authors: | Deshmukh, L, Schwieters, C.D, Grishaev, A, Clore, G.M. | | Deposit date: | 2016-02-05 | | Release date: | 2016-03-30 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Quantitative Characterization of Configurational Space Sampled by HIV-1 Nucleocapsid Using Solution NMR, X-ray Scattering and Protein Engineering.
Chemphyschem, 17, 2016
|
|
6WAP
 
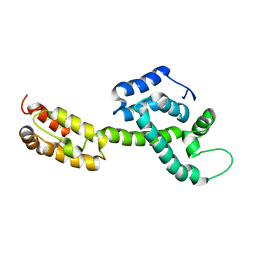 | | Atomic-Resolution Structure of HIV-1 Capsid Tubes by Magic Angle Spinning NMR | | Descriptor: | HIV-1 capsid protein | | Authors: | Lu, M, Russell, R.W, Bryer, A, Quinn, C.M, Hou, G, Zhang, H, Schwieters, C.D, Perilla, J.R, Gronenborn, A.M, Polenova, T. | | Deposit date: | 2020-03-25 | | Release date: | 2020-09-02 | | Last modified: | 2023-06-14 | | Method: | SOLID-STATE NMR | | Cite: | Atomic-resolution structure of HIV-1 capsid tubes by magic-angle spinning NMR.
Nat.Struct.Mol.Biol., 27, 2020
|
|
2E36
 
 | | L11 with SANS refinement | | Descriptor: | 50S ribosomal protein L11 | | Authors: | Lee, D, Walsh, J.D, Yu, P, Markus, M.A, Choli-Papadopoulous, T, Schwieters, C.D, Krueger, S, Draper, D.E, Wang, Y.X. | | Deposit date: | 2006-11-20 | | Release date: | 2007-06-19 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | The structure of free L11 and functional dynamics of L11 in free, L11-rRNA(58 nt) binary and L11-rRNA(58 nt)-thiostrepton ternary complexes
J.Mol.Biol., 367, 2007
|
|
2E35
 
 | | the minimized average structure of L11 with rg refinement | | Descriptor: | 50S ribosomal protein L11 | | Authors: | Lee, D, Walsh, J.D, Yu, P, Markus, M.A, Choli-Papadopoulous, T, Schwieters, C.D, Krueger, S, Draper, D.E, Wang, Y.X. | | Deposit date: | 2006-11-20 | | Release date: | 2007-06-19 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | The structure of free L11 and functional dynamics of L11 in free, L11-rRNA(58 nt) binary and L11-rRNA(58 nt)-thiostrepton ternary complexes
J.Mol.Biol., 367, 2007
|
|
2E34
 
 | | L11 structure with RDC and RG refinement | | Descriptor: | 50S ribosomal protein L11 | | Authors: | Lee, D, Walsh, J.D, Yu, P, Markus, M.A, Choli-Papadopoulous, T, Schwieters, C.D, Krueger, S, Draper, D.E, Wang, Y.X. | | Deposit date: | 2006-11-20 | | Release date: | 2007-06-19 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | The structure of free L11 and functional dynamics of L11 in free, L11-rRNA(58 nt) binary and L11-rRNA(58 nt)-thiostrepton ternary complexes
J.Mol.Biol., 367, 2007
|
|
5T1O
 
 | | Solution-state NMR and SAXS structural ensemble of NPr (1-85) in complex with EIN-Ntr (170-424) | | Descriptor: | Phosphocarrier protein NPr, Phosphoenolpyruvate-protein phosphotransferase PtsP | | Authors: | Strickland, M, Stanley, A.M, Wang, G, Schwieters, C.D, Buchanan, S, Peterkofsky, A, Tjandra, N. | | Deposit date: | 2016-08-19 | | Release date: | 2016-11-16 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Structure of the NPr:EIN(Ntr) Complex: Mechanism for Specificity in Paralogous Phosphotransferase Systems.
Structure, 24, 2016
|
|
2N5T
 
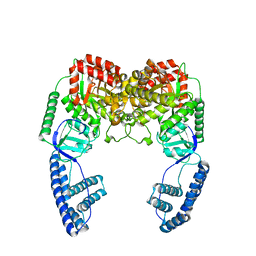 | | Ensemble solution structure of the phosphoenolpyruvate-Enzyme I complex from the bacterial phosphotransferase system | | Descriptor: | Phosphoenolpyruvate-protein phosphotransferase | | Authors: | Venditti, V, Schwieters, C.D, Grishaev, A, Clore, G. | | Deposit date: | 2015-07-28 | | Release date: | 2015-09-02 | | Last modified: | 2019-04-17 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Dynamic equilibrium between closed and partially closed states of the bacterial Enzyme I unveiled by solution NMR and X-ray scattering.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2N9Z
 
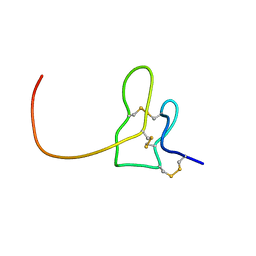 | | Solution structure of K1 lobe of double-knot toxin | | Descriptor: | Tau-theraphotoxin-Hs1a | | Authors: | Bae, C, Anselmi, C, Kalia, J, Jara-Oseguera, A, Schwieters, C.D, Krepkiy, D, Lee, C.W, Kim, E.H, Kim, J.I, Faraldo-Gomez, J.D, Swartz, K.J. | | Deposit date: | 2015-12-16 | | Release date: | 2016-03-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the mechanism of activation of the TRPV1 channel by a membrane-bound tarantula toxin
Elife, 5, 2016
|
|
2N0A
 
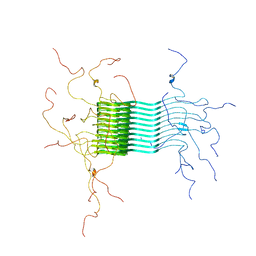 | | Atomic-resolution structure of alpha-synuclein fibrils | | Descriptor: | Alpha-synuclein | | Authors: | Tuttle, M.D, Comellas, G, Nieuwkoop, A.J, Covell, D.J, Berthold, D.A, Kloepper, K.D, Courtney, J.M, Kim, J.K, Schwieters, C.D, Lee, V.M, George, J.M, Rienstra, C.M. | | Deposit date: | 2015-03-04 | | Release date: | 2016-03-23 | | Last modified: | 2023-06-14 | | Method: | SOLID-STATE NMR | | Cite: | Solid-state NMR structure of a pathogenic fibril of full-length human alpha-synuclein.
Nat.Struct.Mol.Biol., 23, 2016
|
|
2NAJ
 
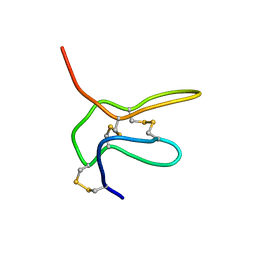 | | Solution structure of K2 lobe of double-knot toxin | | Descriptor: | Tau-theraphotoxin-Hs1a | | Authors: | Bae, C, Anselmi, C, Kalia, J, Jara-Oseguera, A, Schwieters, C.D, Krepkiy, D, Lee, C.W, Kim, E.H, Kim, J.I, Faraldo-Gomez, J.D, Swartz, K.J. | | Deposit date: | 2016-01-04 | | Release date: | 2016-03-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the mechanism of activation of the TRPV1 channel by a membrane-bound tarantula toxin
Elife, 5, 2016
|
|
2N2L
 
 | | NMR structure of yersinia pestis ail (attachment invasion locus) in decylphosphocholine micelles calculated with implicit membrane solvation | | Descriptor: | Outer membrane protein X | | Authors: | Marassi, F.M, Ding, Y, Tian, Y, Schwieters, C.D, Yao, Y. | | Deposit date: | 2015-05-10 | | Release date: | 2015-07-22 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Backbone structure of Yersinia pestis Ail determined in micelles by NMR-restrained simulated annealing with implicit membrane solvation.
J.Biomol.Nmr, 63, 2015
|
|
2P01
 
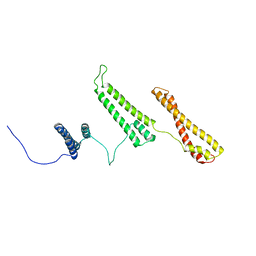 | | The structure of receptor-associated protein(RAP) | | Descriptor: | Alpha-2-macroglobulin receptor-associated protein | | Authors: | Lee, D, Walsh, J.D, Migliorini, M, Yu, P, Cai, T, Schwieters, C.D, Krueger, S, Strickland, D.K, Wang, Y.X. | | Deposit date: | 2007-02-28 | | Release date: | 2007-08-21 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | The structure of receptor-associated protein (RAP).
Protein Sci., 16, 2007
|
|
2P03
 
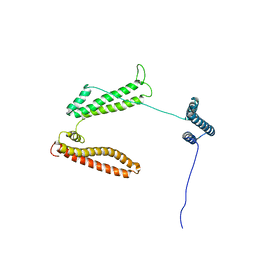 | | The structure of receptor-associated protein(RAP) | | Descriptor: | Alpha-2-macroglobulin receptor-associated protein | | Authors: | Lee, D, Walsh, J.D, Migliorini, M, Yu, P, Cai, T, Schwieters, C.D, Krueger, S, Strickland, D.K, Wang, Y.X. | | Deposit date: | 2007-02-28 | | Release date: | 2007-08-21 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | The structure of receptor-associated protein (RAP).
Protein Sci., 16, 2007
|
|
2L5H
 
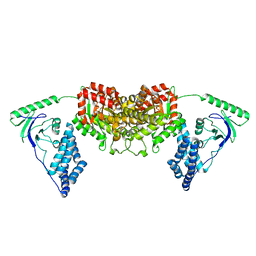 | | Solution Structure of the H189Q mutant of the Enzyme I dimer Using Residual Dipolar Couplings and Small Angle X-Ray Scattering | | Descriptor: | Phosphoenolpyruvate-protein phosphotransferase | | Authors: | Takayama, Y.D, Schwieters, C.D, Grishaev, A, Guirlando, R, Clore, G. | | Deposit date: | 2010-11-01 | | Release date: | 2011-01-12 | | Last modified: | 2012-04-25 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Combined Use of Residual Dipolar Couplings and Solution X-ray Scattering To Rapidly Probe Rigid-Body Conformational Transitions in a Non-phosphorylatable Active-Site Mutant of the 128 kDa Enzyme I Dimer.
J.Am.Chem.Soc., 133, 2011
|
|
2JYH
 
 | |
2JYF
 
 | |
2K4F
 
 | | Mouse CD3epsilon Cytoplasmic Tail | | Descriptor: | T-cell surface glycoprotein CD3 epsilon chain | | Authors: | Xu, C, Call, M.E, Schwieters, C.D, Schnell, J.R, Gagnon, E.E, Wucherpfennig, K.W, Chou, J.J. | | Deposit date: | 2008-06-07 | | Release date: | 2008-12-02 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Regulation of T Cell Receptor Activation by Dynamic Membrane Binding of the CD3varepsilon Cytoplasmic Tyrosine-Based Motif.
Cell(Cambridge,Mass.), 135, 2008
|
|
