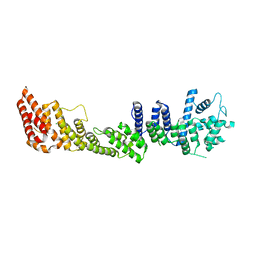
2GED
 
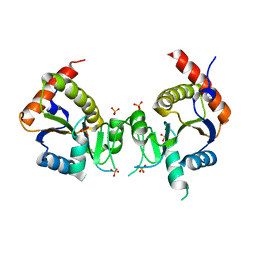 | |
4KF8
 
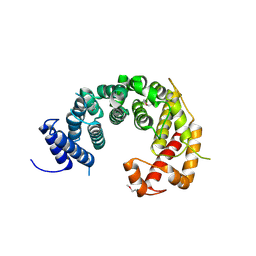 | |
4KF7
 
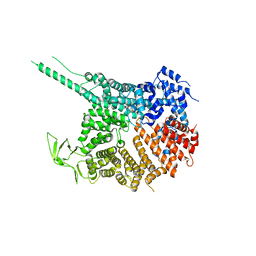 | |
2QX5
 
 | | Structure of nucleoporin Nic96 | | Descriptor: | CHLORIDE ION, Nucleoporin NIC96 | | Authors: | Jeudy, S, Schwartz, T.U. | | Deposit date: | 2007-08-10 | | Release date: | 2007-09-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of nucleoporin Nic96 reveals a novel, intricate helical domain architecture
J.Biol.Chem., 282, 2007
|
|
4TVS
 
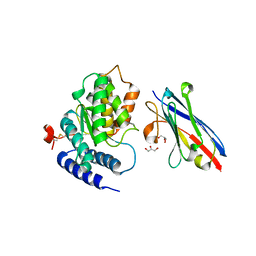 | | LAP1(aa356-583), H.sapiens, bound to VHH-BS1 | | Descriptor: | GLYCEROL, SULFATE ION, Torsin-1A-interacting protein 1, ... | | Authors: | Sosa, B.A, Schwartz, T.U. | | Deposit date: | 2014-06-27 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | How lamina-associated polypeptide 1 (LAP1) activates Torsin.
Elife, 3, 2014
|
|
1XKS
 
 | |
6XF1
 
 | |
6XF2
 
 | |
7TJC
 
 | | VHH Chl-B2 in complex with Chloramphenicol | | Descriptor: | CHLORAMPHENICOL, GLYCEROL, VHH-Chl-B2 | | Authors: | Nordeen, S.A, Schwartz, T.U. | | Deposit date: | 2022-01-16 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure and specificity of an anti-chloramphenicol single domain antibody for detection of amphenicol residues.
Protein Sci., 31, 2022
|
|
3MZK
 
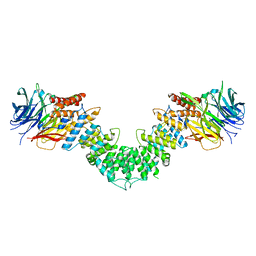 | | Sec13/Sec16 complex, S.cerevisiae | | Descriptor: | Protein transport protein SEC13, Protein transport protein SEC16 | | Authors: | Whittle, J.R, Schwartz, T.U. | | Deposit date: | 2010-05-12 | | Release date: | 2010-08-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structure of the Sec13-Sec16 edge element, a template for assembly of the COPII vesicle coat.
J.Cell Biol., 190, 2010
|
|
3MZL
 
 | | Sec13/Sec31 edge element, loop deletion mutant | | Descriptor: | Protein transport protein SEC13, Protein transport protein SEC31 | | Authors: | Whittle, J.R, Schwartz, T.U. | | Deposit date: | 2010-05-12 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Sec13-Sec16 edge element, a template for assembly of the COPII vesicle coat.
J.Cell Biol., 190, 2010
|
|
5DJ4
 
 | |
5T0N
 
 | |
4DXR
 
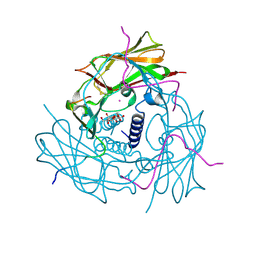 | | Human SUN2-KASH1 complex | | Descriptor: | DECYL-BETA-D-MALTOPYRANOSIDE, Nesprin-1, POTASSIUM ION, ... | | Authors: | Sosa, B, Schwartz, T.U. | | Deposit date: | 2012-02-28 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | LINC Complexes Form by Binding of Three KASH Peptides to Domain Interfaces of Trimeric SUN Proteins.
Cell(Cambridge,Mass.), 149, 2012
|
|
4DXT
 
 | | Human SUN2 (AA 522-717) | | Descriptor: | POTASSIUM ION, SUN domain-containing protein 2 | | Authors: | Sosa, B, Schwartz, T.U. | | Deposit date: | 2012-02-28 | | Release date: | 2012-06-06 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | LINC Complexes Form by Binding of Three KASH Peptides to Domain Interfaces of Trimeric SUN Proteins.
Cell(Cambridge,Mass.), 149, 2012
|
|
4DXS
 
 | | Human SUN2-KASH2 complex | | Descriptor: | DECYL-BETA-D-MALTOPYRANOSIDE, Nesprin-2, POTASSIUM ION, ... | | Authors: | Sosa, B, Schwartz, T.U. | | Deposit date: | 2012-02-28 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | LINC Complexes Form by Binding of Three KASH Peptides to Domain Interfaces of Trimeric SUN Proteins.
Cell(Cambridge,Mass.), 149, 2012
|
|
7PER
 
 | | Model of the inner ring of the human nuclear pore complex | | Descriptor: | Nuclear pore complex protein Nup155, Nuclear pore complex protein Nup205, Nuclear pore complex protein Nup93, ... | | Authors: | Schuller, A.P, Wojtynek, M, Mankus, D, Tatli, M, Kronenberg-Tenga, R, Regmi, S.G, Dasso, M, Weis, K, Medalia, O, Schwartz, T.U. | | Deposit date: | 2021-08-11 | | Release date: | 2021-10-20 | | Last modified: | 2021-11-24 | | Method: | ELECTRON MICROSCOPY (35 Å) | | Cite: | The cellular environment shapes the nuclear pore complex architecture.
Nature, 598, 2021
|
|
7PEQ
 
 | | Model of the outer rings of the human nuclear pore complex | | Descriptor: | Nuclear pore complex protein Nup107, Nuclear pore complex protein Nup133, Nuclear pore complex protein Nup160, ... | | Authors: | Schuller, A.P, Wojtynek, M, Mankus, D, Tatli, M, Kronenberg-Tenga, R, Regmi, S.G, Dasso, M, Weis, K, Medalia, O, Schwartz, T.U. | | Deposit date: | 2021-08-11 | | Release date: | 2021-10-20 | | Last modified: | 2021-11-24 | | Method: | ELECTRON MICROSCOPY (35 Å) | | Cite: | The cellular environment shapes the nuclear pore complex architecture.
Nature, 598, 2021
|
|
3CQC
 
 | | Nucleoporin Nup107/Nup133 interaction complex | | Descriptor: | Nuclear pore complex protein Nup107, Nuclear pore complex protein Nup133 | | Authors: | Jeudy, S, Boehmer, T, Berke, I, Schwartz, T.U. | | Deposit date: | 2008-04-02 | | Release date: | 2008-07-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural and functional studies of Nup107/Nup133 interaction and its implications for the architecture of the nuclear pore complex.
Mol.Cell, 30, 2008
|
|
3CQG
 
 | | Nucleoporin Nup107/Nup133 interaction complex, delta finger mutant | | Descriptor: | Nuclear pore complex protein Nup107, Nuclear pore complex protein Nup133 | | Authors: | Jeudy, S, Boehmer, T, Berke, I, Schwartz, T.U. | | Deposit date: | 2008-04-02 | | Release date: | 2008-07-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and functional studies of Nup107/Nup133 interaction and its implications for the architecture of the nuclear pore complex.
Mol.Cell, 30, 2008
|
|
3EWE
 
 | | Crystal Structure of the Nup85/Seh1 Complex | | Descriptor: | Nucleoporin NUP85, Nucleoporin SEH1 | | Authors: | Brohawn, S.G, Leksa, N.C, Rajashankar, K.R, Schwartz, T.U. | | Deposit date: | 2008-10-14 | | Release date: | 2008-11-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural evidence for common ancestry of the nuclear pore complex and vesicle coats.
Science, 322, 2008
|
|
6OIF
 
 | | Cryo-EM structure of human TorsinA filament | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Torsin-1A | | Authors: | Zheng, W, Demircioglu, F.E, Schwartz, T.U, Egelman, E.H. | | Deposit date: | 2019-04-09 | | Release date: | 2019-07-24 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | The AAA + ATPase TorsinA polymerizes into hollow helical tubes with 8.5 subunits per turn.
Nat Commun, 10, 2019
|
|
4YD8
 
 | |
4YGA
 
 | |
4YCZ
 
 | |
